Microbiome studies have traditionally fallen into studies of who’s there and what are they doing. To address these questions, biologists often use next-generation sequencing. Sequencing the 16S rRNA reveals the identity of the organisms present while sequencing of all transcripts gives clues into what the microbes are doing.
But aside from sequencing, scientists can also study the microbiome by using engineered genetic tools and reporter microbes. Let’s take a look at a few of these methods below.
Engineering gut microbes in situ
While the ability to monitor the microbiome with sequencing is vast, the ability to genetically alter microbiome members is limited and many microbes are difficult to culture outside of their natural environment. Thus, the Harris Wang lab at Columbia University developed a method for genetically altering gut microbiome members in their native habitat (Ronda et al., 2019). They called their method Metagenomic Alteration of Gut microbiome by In situ Conjugation (MAGIC).
With MAGIC, scientists introduce an Escherichia coli donor strain into the microbiome to deliver a genetic payload to the recipients using the IncP⍺-family-RP4 conjugation system. This system can deliver plasmids to both Gram-positive and Gram-negative cells.
The Wang lab further developed a modular suite of plasmids (pGT) with various replicative origins that can be used for broad or narrow range of microbial species. These also contain the RP4 transfer origin as well as a selectable marker and a desired genetic payload (ex: GFP). Thus, cells can be both sorted using FACS and isolated with antibiotic selection.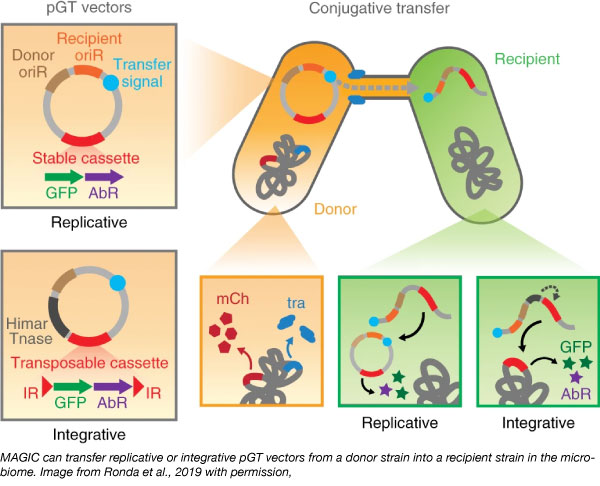
The team tested this system in vitro and in vivo. They identified cells from 19 genera across four phyla that have taken up the plasmid through conjugation in vivo studies. However, they only identified seven genera in in vitro experiments, possibly because in vitro conditions did not optimally support growth of diverse species. These results further highlight the need for in situ genetic tools for microbiome studies.
Paper-based synthetic biology approach to analyze gut microbiota and host biomarkers
In clinical and diagnostic labs, deep sequencing and qPCR methods are too costly and time intensive to process a large number of microbiome samples. To enable large-scale, longitudinal studies of the gut microbiome, James Collins’s lab developed a paper-based method that can be used to detect microbes and biomarkers on paper.
At the heart of their method is the RNA toehold switch, a RNA hairpin that can be designed to bind and detect nearly any RNA sequence. Downstream of the hairpin is a ribosome binding site (RBS) followed by a reporter gene. The hairpin blocks translation by sequestering the RBS, but binding of the “trigger RNA” (in this case, biomarker RNA or 16S rRNA isolated from the microbial community) reveals the RBS and allows translation of the reporter gene resulting in GFP signal. With the addition of standards, the assay is semi-quantitative and can approximate mRNA concentration within a sample.
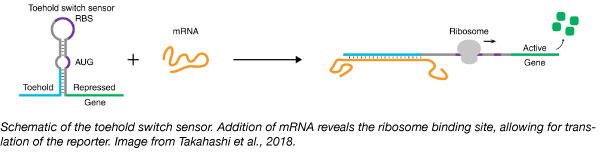 Using this system, the Collins lab was able to detect mRNA from 10 bacteria of clinical significance and four clinically relevant host biomarkers.
Using this system, the Collins lab was able to detect mRNA from 10 bacteria of clinical significance and four clinically relevant host biomarkers.
The lab validated their platform on clinical stool samples by comparison to qPCR and demonstrated that it can also detect expression of toxin mRNA from Clostridium difficile. As mRNA expression cannot be identified with DNA-based qPCR methods, the ability to detect mRNA transcripts of toxins and other transcripts of interest is a benefit of this method.
Find the toehold switch microbiota/biomarker sensors here
Studying the bee microbiome using the Bee Microbiome Toolkit (BTK)
Another system that lacks genetic tools is the bee microbiome. Nancy Moran’s and Jeffrey Barrick’s labs at The University of Texas at Austin recently developed the Bee Microbiome Toolkit (BTK) to study bumble bee and honey bee microbiome. The toolkit consists of modular parts that are assembled with Golden Gate Assembly.
They built this toolkit upon the Yeast Toolkit by replacing the backbone vector’s high-copy number bacterial ColE1 origin with the RSF1010 origin, an IncQ origin with broad host-range (Meyer, 2009). The vector retains the oriT for delivery into recipients by conjugation.
The labs screened for the host-range of the plasmids and suitability of various antibiotic resistance markers, promoters, and inducible gene expression systems in bee gut microbiome species such as Snodgrassella alvi and Gilliamella apicoli. The plasmids could also be transformed into Alphaproteobacteira, Betaproteobacteria, and Gammaproteobacteria species isolated from different bee species.
BTK can be used to introduce fluorescent markers to visualize localization and distribution of gut bacteria without fixation or preparation, and deliver CRISPR components for gene disruption or CRISPRi. While designed to study the bee microbiome, these broad-host-range plasmid could also be used to study other microbiome systems.
Find the toolkit for modular assembly of broad host range plasmids
Counting cell division in the gut microbiota
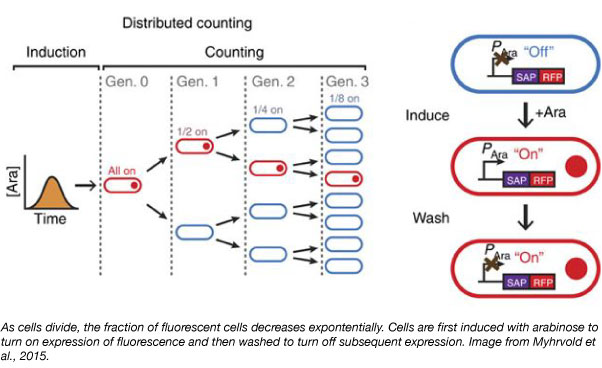
Monitoring microbial population dynamics in the gut can provide insight into how the microbiome responds to infection, dysbiosis, and antibiotic treatment. To facilitate these studies, Pamela Silver’s lab developed a synthetic mark and recapture strategy for bacteria to count cell divisions. The technique, called distributed cell division counting (DCDC), relies on the assembly of fluorescent proteins into one bright particle that segregates into one cell during cell division.
To achieve the agglomeration of fluorescent proteins into one particle within the cell, the lab fused the red fluorescent protein (RFP) to a self-assembling protein (SAP) such as a bacteriophage shell protein or a bacterial microcompartment protein. The expression of this fusion is under control of the arabinose-inducible promoter.
Cells are first induced with arabinose to begin expressing the SAP-RFP fusion. Then, the cells are washed to stop the production of new particles. With subsequent cell divisions, the fraction of cells containing the particle exponentially decreases over time and the number of elapsed generations can be determined. The lab used DCDC to measure growth rate of E. coli for over 10 generations and showed that this method can be used for both in vitro and in vivo studies.
Find distributed cell division counting plasmids here
References
Leonard, Sean P., et al. "Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids." ACS synthetic biology 7.5 (2018): 1279-1290. PubMed PMID: 29608282. PubMed Central PMCID: PMC5963681.
Meyer, Richard. "Replication and conjugative mobilization of broad host-range IncQ plasmids." Plasmid 62.2 (2009): 57-70. PubMed PMID: 19465049. PubMed Central PMCID: PMC2752045.
Myhrvold, Cameron, et al. "A distributed cell division counter reveals growth dynamics in the gut microbiota." Nature communications 6 (2015): 10039. PubMed PMID: 26615910. PubMed Central PMCID: PMC4674677.
Ronda, Carlotta, et al. "Metagenomic engineering of the mammalian gut microbiome in situ." Nature methods 16.2 (2019): 167. PubMed PMID: 30643213. PubMed Central PMCID: PMC6467691.
Takahashi, Melissa K., et al. "A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers." Nature communications 9.1 (2018): 3347. PubMed PMID: 30131493. PubMed Central PMCID: PMC6104080.
Resources on the Addgene Blog
- Find microbiology blog posts
- Read about synthetic biology on the blog
- Find all of our Plasmids 101 blog posts
Resources on Addgene.org
- Check out our microbiology resource page
- Deposit microbiology plasmids with Addgene
- Find synthetic biology plasmids
Topics: Microbiology, Other Plasmid Tools, Plasmids





Leave a Comment