In its simplest form, PCR based cloning is about making a copy of a piece of DNA and at the same time adding restriction sites to the ends of that piece of DNA so that it can be easily cloned into a plasmid of interest. You can use similar processes to add overhangs to your insert of interest for Gibson assembly. The steps following primer design and the PCR process itself are very similar to those outlined in our restriction cloning post with a few quirks specific to the PCR cloning process - please check out that post if you need a more detailed refresher on the downstream steps.
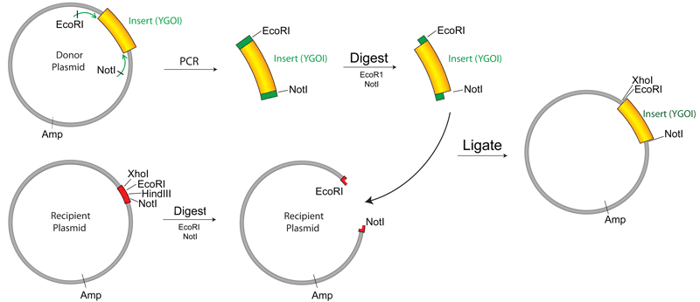
For this example, we will describe how to copy a cDNA from one vector into a new vector that is better suited for analyzing the gene’s function. The process is shown graphically in the following figure, in which we add EcoRI and NotI sites to Your Gene of Interest (YGOI) for ligation into a recipient plasmid.
Designing primers for PCR based cloning
The basic PCR primers for molecular cloning consist of:
-
Leader Sequence: Extra base pairs on the 5' end of the primer assist with restriction enzyme digestion (usually 3-6bp)
-
Restriction Site: Your chosen restriction site for cloning (usually 6-8bp)
-
Hybridization Sequence: The region of the primer that binds to the sequence to be amplified (usually 18-21bp)
When selecting restriction sites, you should use a DNA analysis tool, such as Addgene’s Sequence Analyzer, to allow you to identify which restriction sites are present in a given sequence. You want to choose enzymes that:
-
Do not cut within your insert
-
Are in the desired location in your recipient plasmid (usually in the Multiple Cloning Site (MCS)), but do not cut elsewhere on the plasmid
-
Bonus: It is helpful to choose restriction enzymes that can both function in the same buffer, as this will save time later
In our example, we will use EcoRI and NotI to ligate our cDNA into the recipient plasmid. Remember to insert your DNA in the correct orientation in the recipient plasmid by viewing the MCS and fusing the upstream restriction site to the forward primer and the downstream restriction site to the reverse primer.
Next, we need to examine the DNA sequence that we want to amplify and design primers that will bind to and replicate it. The following image shows the ends of the ORF and how these are used for primer design:
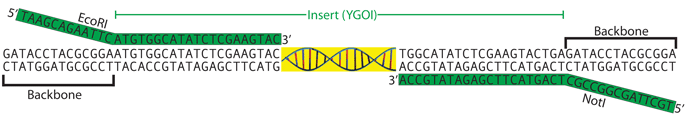
Because we are cloning an ORF, we want to clone from the start codon (ATG) to the stop codon (TGA, in this example). Assuming you are amplifying from plasmid DNA (rather than from genomic DNA or a cDNA library), roughly 18-21bp is usually sufficient to give specificity and to also be compatible with a standard PCR reaction (see PCR Video). Therefore, our Forward Primer will use the sequence 5'-ATGTGGCATATCTCGAAGTAC-3' for the region that binds the ORF and we will add the EcoRI restriction site (GAATTC) to the 5’ end of this primer, making our Forward Primer 5'-GAATTCATGTGGCATATCTCGAAGTAC-3'.
Many restriction enzymes do not cut DNA efficiently at the end of a linear piece (see NEB for more information). Thus, we recommend that you add 3-6 bases upstream of your restriction site to improve cutting efficiency. You can generally add any 6 bases, but you should ensure that the bases do not result in the formation of a hairpin structure within your primer. In our case, we will add TAAGCA, resulting in a final Forward Primer sequence of 5'-TAAGCAGAATTCATGTGGCATATCTCGAAGTAC-3'.
For the Reverse Primer, the design is similar, but we need to use the reverse complement to get PCR amplification. We can start similarly, taking the final 18 bases of the ORF, including the stop codon (5'-TGGCATATCTCGAAGTACTGA-3'), then adding NotI (GCGGCCGC) and then TAAGCA to improve restriction enzyme digestion. This gives us a sequence of 5'-TGGCATATCTCGAAGTACTGAGCGGCCGCTAAGCA-3' (30bp with 18bp of homology to the ORF). We now need to generate the reverse-complement of this sequence so that we can successfully amplify the ORF. You can generate the reverse-complement using existing software (a quick internet search will lead you to here and many others). If we put the sequence we chose for our reverse primer (5’-TGGCATATCTCGAAGTACTGAGCGGCCGCTAAGCA-3’) into this calculator we get a final Reverse Primer sequence of 5’-TGCTTAGCGGCCGCTCAGTACTTCGAGATATGCCA-3’.
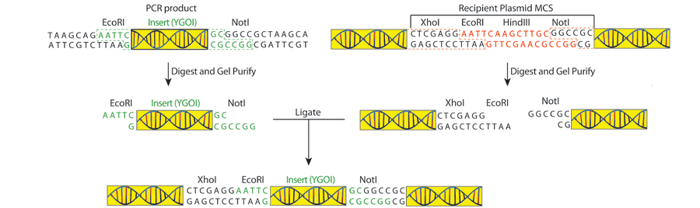
Preparing the PCR product for cloning
Run the PCR reaction
Run PCR to amplify your insert DNA. It is important to use a high fidelity polymerase to minimize mutations. The fidelity of the polymerase becomes more important the longer the expected PCR product is. You should select an annealing temperature based on the melting temperature (Tm) of the portion of the primer that hybridizes to the sequence to be amplified (the ORF in this case), not the Tm of the entire primer. If you are amplifying from a plasmid or simple template, there is very little chance for mis-priming, so you can use a pretty wide range of annealing temperatures, but you may need to increase your primer length and increase the Tm if you are trying to clone from genomic DNA, a cDNA library, or by RT-PCR.
Isolate the PCR product
Isolate your PCR product from the rest of the PCR reaction using a kit, such as the QIAquick PCR Purification Kit. The PCR product is now ready for restriction digestion. As such, the later steps in this process are the same as those discussed in our restriction cloning post. However, even more than with standard restriction cloning, it is essential that you verify your final plasmid by sanger sequencing. DNA replication by PCR has error rates that range from roughly 1 per 500bp to roughly 1 per 10 million bp depending on the polymerase used. No matter which polymerase you use, it is important that you sequence the final product.
Additional Resources on the Addgene Blog
- Learn About Other Cloning Techniques
- Catch Up on All Things Plasmid with Our Plasmids 101 Series
- Get Tips on Verifying Your Plasmid
Resources on Addgene.org
Topics: Plasmid Cloning, PCR, Plasmids






Leave a Comment