Working with AAV vectors in the brain and peripheral nervous system presents a special challenge to scientists. What works well in one cell or neuron type may not work well for a neighboring cell type, even within the same brain region or nucleus. Just optimizing AAV expression can take months (or longer), yet the process is often not detailed in the primary literature, and as a result can leave scientists under-informed with respect to how different serotypes, promoters, and injection methods are best suited to their particular scientific question - even when others have already answered this question and used it to publish!
Working with AAV vectors in neuroscience
Let’s consider my first challenge when I started working with AAV vectors. I wanted to use an AAV vector to express ChR2(H134R) in A2 norepinephrine expressing neurons. We tested four serotypes containing an expression cassette with an ubiquitous promoter to express GFP in a group of young rats. We found that, in this case, AAV1 (23%) and AAV6 (<5%, data not shown) were not good options while AAV8 (82%) and AAV9 (84%) were both excellent at infecting our target neurons (Figure 1). However, this one set of experiments didn’t provide any information on other important factors such as what is the best promoter to use, or if and how much the spread of the virus to adjacent areas impacted the experiments we wanted to perform. It required a great deal of time and effort to test and optimize our vector - overall, I spent two years testing and reformulating our AAV construct to optimize performance in our system and yet none of that data was published.
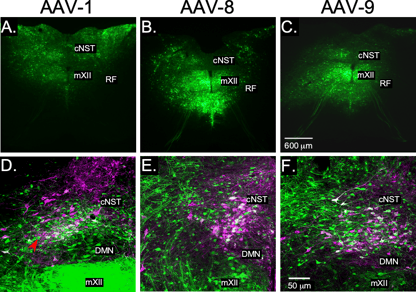 |
| Fig. 1: Testing of three different AAV serotypes (AAV1, 8, 9) injected into the A2 noradrenergic area of the caudal NST. (A-C) Low-magnification images showing viral spread. (D-F) High-magnification images immuno-stained against dopamine beta-hydroxylase (DBH, magenta) to label norepinephrine (NE) expressing neurons and GFP (green) to label neurons transduced by each serotype. |
Working at Addgene has shown me this was far from an isolated case. Our scientific support team answers hundreds of AAV questions every year on a range of issues related to AAV vectors, many from people trying to answer the exact same question I was: What AAV vector is best for my specific system?
Sharing data and community concerns
AAV vectors are complex biological reagents; many factors, like storage buffers and conditions (3,5,8), injection titers (1,2), and cell types used for packaging viral particles (6), can influence vector performance. Open science initiatives, like Addgene’s Data Hub, provide an avenue to more easily solve these frustrating and often months-long quests to find the right AAV for your system or to successfully replicate published works. Of particular importance to these efforts is the open sharing of negative data.
Negative AAV data is highly desirable
In a survey we conducted to improve our Addgene Data Hub, 76% of respondents indicated they wanted to see more negative data (Figure 2.) At the same time, when scientists who support open science, but do not share their own data - which seems to be the majority of scientists (4,7,9) - are asked why they don’t share, many express concern about sharing negative data they cannot explain, perhaps out of fear it points to flaws or gaps in their technical ability. But, especially for AAV data, that may not be the case.
We can all acknowledge that negative data is something every researcher experiences and that it often provides incredibly valuable insight into one’s work. As a community, we must therefore actively work to remove the stigma around sharing negative data and instead focus on getting this information out into the public where it can be utilized by others. Addgene’s AAV Data Hub is one resource where this type of data is not only housed, but encouraged.
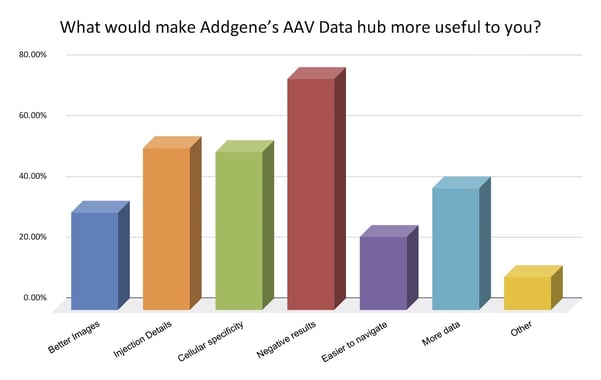 |
| Fig. 2: Results from a survey conducted by Addgene in 2021 asking scientists what data would improve our AAV Data Hub. |
The Addgene Data Hub is here to help!
The Addgene Data Hub is designed to help scientists understand how our vectors perform in various experimental settings. It is a curated set of technical reports on AAV vectors from Addgene and the CLOVER Center, each one submitted by researchers like you. Reports are reviewed by Addgene scientists for clarity and accuracy, and by the end of the year, those that meet our minimum standards (clear and detailed methods and image descriptions, a minimum n = 2, and well labeled high- and low-magnification images demonstrating vector performance) will be assigned citable DOIs through DataCite before being made publicly available through the Data Hub. These minimum data standards are designed specifically to allow scientists to evaluate how small changes in vector administration can influence AAV expression properties.
By providing many of the details not generally captured in the literature, scientists can use the Data Hub to compare different methods and vectors, and use this data to guide them in their own experiments. By providing DOIs, this information can be cited, tracked, and appropriately credited, broadening the knowledge base of AAV vector performance in various brain regions and cell types.
Power in numbers
A Pubmed search for “AAV” and “brain” over the past decade gives over 22,000 results - an enormous amount of data potentially available to evaluate AAV vector performance. Yet, most of the technical data needed to replicate and build on others’ results is available not through published manuscripts, but through word of mouth or old lab notebooks and computer files. If you asked your team members or colleagues: How much unpublished AAV data do you have sitting in computers and notebooks right now?, I’m willing to bet the answer would be along the lines of “we have a lot”. In fact, scientists often cite “word of mouth” as their source for this type of technical know-how, which highlights the dearth of AAV vector information provided in openly accessible forms like publications. Why not get this data out there?
The Addgene Data Hub is a free resource to share this data and help speed scientific progress, but it will only be effective if many scientists choose to help each other by sharing their data. So take advantage of it and get your data - negative AND positive - out into the public eye where it can benefit the community. We are all in this together and everybody, helping everybody, helps everybody!
References and resources
References
- Dayton, R. D., Grames, M. S., & Klein, R. L. (2018). More expansive gene transfer to the rat CNS: AAV PHP.EB vector dose–response and comparison to AAV PHP.B. Gene Therapy, 25(5), 392–400. https://doi.org/10.1038/s41434-018-0028-5
- de Backer, M. W. A., Brans, M. A. D., Luijendijk, M. C., Garner, K. M., & Adan, R. A. H. (2010). Optimization of Adeno-Associated Viral Vector-Mediated Gene Delivery to the Hypothalamus. Human Gene Therapy, 21(6), 673–682. https://doi.org/10.1089/hum.2009.169
- Gruntman, A. M., Su, L., Su, Q., Gao, G., Mueller, C., & Flotte, T. R. (2015). Stability and compatibility of recombinant adeno-associated virus under conditions commonly encountered in human gene therapy trials. Human Gene Therapy Methods, 26(2), 71–76. https://doi.org/10.1089/hgtb.2015.040
- Initiative, F. B. (2019). Setting your data free. Nature, 5–7.
- O’Connor, D. M., Lutomski, C., Jarrold, M. F., Boulis, N. M., & Donsante, A. (2019). Lot-to-Lot Variation in Adeno-Associated Virus Serotype 9 (AAV9) Preparations. Human Gene Therapy Methods, 30(6), 214–225. https://doi.org/10.1089/hgtb.2019.105
- Rumachik, N. G., Malaker, S. A., Poweleit, N., Maynard, L. H., Adams, C. M., Leib, R. D., Cirolia, G., Thomas, D., Stamnes, S., Holt, K., Sinn, P., May, A. P., & Paulk, N. K. (2020). Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors. Molecular Therapy - Methods and Clinical Development, 18(September), 98–118. https://doi.org/10.1016/j.omtm.2020.05.018
- Tedersoo, L., Küngas, R., Oras, E., Köster, K., Eenmaa, H., Leijen, Ä., Pedaste, M., Raju, M., Astapova, A., Lukner, H., Kogermann, K., & Sepp, T. (2021). Data sharing practices and data availability upon request differ across scientific disciplines. Scientific Data, 8(1), 1–11. https://doi.org/10.1038/s41597-021-00981-0
- Wright, J. F., Le, T., Prado, J., Bahr-Davidson, J., Smith, P. H., Zhen, Z., Sommer, J. M., Pierce, G. F., & Qu, G. (2005). Identification of factors that contribute to recombinant AAV2 particle aggregation and methods to prevent its occurrence during vector purification and formulation. Molecular Therapy, 12(1), 171–178. https://doi.org/10.1016/j.ymthe.2005.02.021
- Zuk, P., Sanchez, C. E., Kostick, K., Torgerson, L., Muñoz, K. A., Hsu, R., Kalwani, L., Sierra-Mercado, D., Robinson, J. O., Outram, S., Koenig, B. A., Pereira, S., McGuire, A. L., & Lázaro-Muñoz, G. (2020). Researcher Perspectives on Data Sharing in Deep Brain Stimulation. Frontiers in Human Neuroscience, 14(December), 1–12. https://doi.org/10.3389/fnhum.2020.578687
Additional Resources on addgene.org
Download Addgene's Viral Vectors 101 e-book
Additional Resources on the Addgene Blog
Find and Share AAV Data with Addgene's New AAV Data Hub
Celebrate Open Data Day with Addgene’s AAV Data Hub!
Data Freedom: The Expansion of Data Sharing in Research Publications
Topics: Open Science, Adenoviral Vectors, Addgene’s Viral Service, Data Hub







Leave a Comment