Mice are a common model organism used to understand mammalian traits and genetically engineered mouse models provide researchers with useful and adaptable tools to perform basic and preclinical research. For scientists new to using mouse models, the possibilities may seem endless - and overwhelming.
In the first blog post in this series, I’ll highlight terminology you should be familiar with before working with mouse models, several common techniques used to create engineered mouse models at embryonic stages, and the pros and cons of different genome editing techniques.
Inbred and outbred strains
Mouse strains used in research can fall under one of two broad classifications: inbred or outbred. Inbred strains share a common genetic background, which means that each mouse is genetically identical and all mice of the strain are homozygous at nearly all loci. This reduces variability in genotype and phenotype and helps improve the reproducibility of experiments. Inbred mice are particularly useful for understanding a specific effect related to an experimental manipulation. Common inbred strains include C57BL/6, Balb/c, and 129.
Conversely, outbred strains were originally derived from random crosses between mice of uncharacterized backgrounds to create a limited but genetically diverse gene pool. Outbred strains are strategically bred to maintain maximum genetic heterozygosity in the population, meaning they more closely mimic the genetic diversity of the human population. Outbred strains of mice are useful for comparing dominant and recessive traits and their impact on diseases or treatments, but are not useful for genetic engineering. Examples of outbred strains include Swiss Webster and Harlan (Hsd) National Institutes of Health (NIH) Swiss.
Genetically engineered mice
Mice can be engineered by inserting a targeting vector containing the gene of interest into a zygote, or into embryonic stem cells that are then injected into a blastocyst. There are several types of genetically engineered mice: transgenic mice, knockout mice, and mice with conditional or inducible gene expression.
Transgenic and knockin mice
As the name suggests, transgenic and knockin mice contain an additional gene incorporated into their genome. While transgenic mice have the new gene construct incorporated randomly into the genome, knockin mice have the gene construct inserted at a specific site.
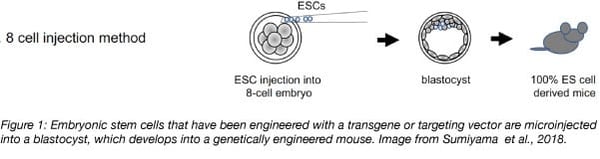
To generate a transgenic mouse, microinjection of DNA or infection with a viral vector containing the gene construct can be used to insert the gene of interest into a zygote or into embryonic stem cells. Non-homologous recombination incorporates this DNA randomly into the mouse genome. In contrast, knockin strains are generated via homologous recombination. A targeting vector is electroporated into embryonic stem cells and this will insert the gene at a specific site within the genome using homology arms that flank the gene of interest. In either case, a positive selection marker such as an antibiotic resistance gene is also included to select for successfully modified zygotes or embryonic stem cells. Modified embryonic stem cells are injected into a blastocyst which is then implanted into a female animal for gestation.
 Nucleases that induce double-stranded DNA breaks, such as ZFN and TALEN, can used to engineer knockin mice. Co-injection of a targeting vector containing homologous ends to the breakpoint promotes homologous recombination of the targeting vector. CRISPR/Cas9-based systems have been developed for generating knockin mice but because CRISPR/Cas9 engineering favors DNA repair via non-homologous end joining, the newly inserted gene may have errors introduced to the nucleotide sequence during the DNA repair process. The addition of small molecules that promote homologous recombination or the use of long single-stranded DNA have led to a higher efficiency of homologous recombination for precise gene insertion. However, CRISPR/Cas9 systems are more useful for generating knockout mice.
Nucleases that induce double-stranded DNA breaks, such as ZFN and TALEN, can used to engineer knockin mice. Co-injection of a targeting vector containing homologous ends to the breakpoint promotes homologous recombination of the targeting vector. CRISPR/Cas9-based systems have been developed for generating knockin mice but because CRISPR/Cas9 engineering favors DNA repair via non-homologous end joining, the newly inserted gene may have errors introduced to the nucleotide sequence during the DNA repair process. The addition of small molecules that promote homologous recombination or the use of long single-stranded DNA have led to a higher efficiency of homologous recombination for precise gene insertion. However, CRISPR/Cas9 systems are more useful for generating knockout mice.
Knockout mice
Knockout mice have undergone genetic modification that alters or eliminates the expression of a particular gene. Targeting vectors used to create knockout mice include a reporter gene, which replaces the targeted gene and provides a way to track successfully engineered cells. Similar to transgenic mice, knockout mice are engineered during the embryonic stem cell stage using one of several techniques. The longest standing technique is the injection of a targeting vector into embryonic stem cells, which are then injected into a blastocyst.
Nucleases can also be used to engineer knockout mice. ZFN, TALEN, and CRISPR/Cas9 systems can all be targeted to specific regions of the genome, and induce non-homologous repair at the site of DNA damage. These nucleases are microinjected into a zygote, which is then implanted into a female mouse for gestation. All three can introduce multiple base pair deletions, but CRISPR/Cas9 can also introduce point mutations.
While using targeted nucleases to generate an engineered mouse model has advantages over the injection of a targeting vector alone, some challenges still exist. Nucleases may cause off-target effects on regions of the genome with similar homology to the site of interest or create mosaic animals where not all cells in the blastocyst were successfully edited by the injection of embryonic stem cells. Also, because non-homologous repair is a random process, zygotes edited using the same nuclease may not result in genetically identical mice. Thus, scientists must genotype each animal to identify the ones with the desired genotype.
Inducible and conditional systems
Conditional knockouts or knockins
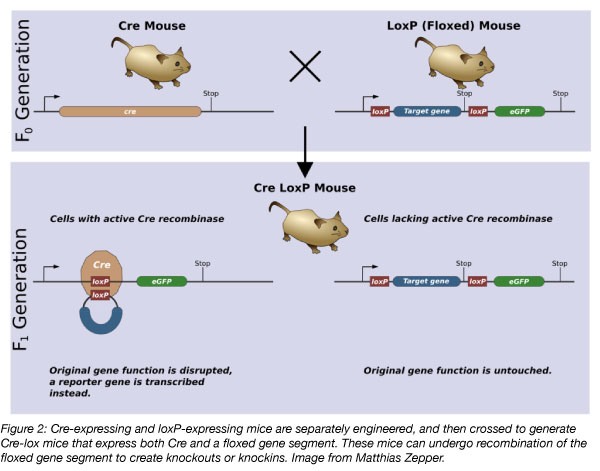
A subset of transgenic mice, conditional knockout/knockin mice, use the Cre-lox recombination system to express the transgene in specific cells or under certain stimuli. Cre recombinase catalyzes the recombination of DNA sequences located between two loxP sites. The loxP sites are present at both ends of a gene in the targeting vector, creating the floxed gene segment. Crossing of mice containing a floxed gene to a Cre-expressing strain (which will be covered in more detail in part 2 of this blog series) controls the cell types in which the conditional recombination can occur based on the cells in which Cre is expressed.
Several variations of this technology can be used to customize the effects of this recombination. To generate a conditional knockout mouse, loxP sites are inserted directly into the genome to flank the gene that will be deleted following Cre-mediated recombination. In conditional knockin mice, the target gene is initially inhibited by the presence of a stop cassette upstream of the target gene in the targeting vector. The stop cassette is floxed, so Cre-mediated recombination deletes the stop cassette, which permits expression of the target gene. In addition, Cre-lox systems can be used to invert a segment of DNA or to translocate a piece of DNA from one region of the genome to another.
Inducible gene expression
Another type of genetically engineered mouse model allows gene expression to be controlled through a drug or small molecule that regulates Cre expression. The most common drug used to induce the expression of Cre is tamoxifen. In tamoxifen-inducible Cre (Cre-ER) systems, Cre is fused to the ligand binding domain of estrogen receptor alpha (Esr1). Mice are fed or injected with tamoxifen, which relieves natural inhibition of the Esr1 domain that blocks Cre expression. Then, Cre is expressed and Cre-mediated recombination of floxed sites in the cell can occur. Similar systems include tetracycline or doxycycline mediated activation of Cre to either induce or repress Cre expression using TetON or TetOFF systems.
So now hopefully creating a genetically engineered mouse seems less overwhelming! In the second blog post of this series, I’ll discuss crossing and maintaining a mouse strain.
Additional resources on the Addgene blog
- Find our CRISPR blog posts
- Read blog posts about using Cre-lox
- Read our genome engineering blog posts
Resources on Addgene.org
- Find more about Cre-lox
- Read our Genome Engineering Guide
- Learn more in our CRIPSR guide






Leave a Comment