 Last Wednesday we worked with the Harvard GSAS Science Policy Group to organize a Minisymposium on Reproducibility. The minisymposium focused on solutions to reproducibility issues in the biological sciences and featured speakers from academia, industry, nonprofits, and publishing. The livestream video from the event can be found below along with a description of the program beneath it. You can jump to the different time stamps in the description to watch any sections you’re particularly interested in, but I’d recommend watching the whole livestream for a more holistic understand of reproducibility issues and their potential solutions.
Last Wednesday we worked with the Harvard GSAS Science Policy Group to organize a Minisymposium on Reproducibility. The minisymposium focused on solutions to reproducibility issues in the biological sciences and featured speakers from academia, industry, nonprofits, and publishing. The livestream video from the event can be found below along with a description of the program beneath it. You can jump to the different time stamps in the description to watch any sections you’re particularly interested in, but I’d recommend watching the whole livestream for a more holistic understand of reproducibility issues and their potential solutions.
Prior to this event, I gave my own talk on reproducibility at Addgene and here I summarize what I learned both in preparation for my talk and at the minisymposium. You can find a variety of additional resources and information about organizations promoting reproducibility in this booklet (which was also handed out at the event).
Program
- 0 - 1:24 - Intro
- 1:24 - 29:31 - Reproducibility Overview - Jeffrey S. Flier, Researcher at Harvard Medical School, former
dean of the faculty of Medicine at Harvard University - 29:31 - 49:45 - Reagent Sharing - Susanna Bachle, Addgene the nonprofit plasmid repository
- 49:45 0 1:18:45 - Reagent Development - Steven C. Almo, Institute for Protein Innovation
- 1:18:45 - 2:01:18 (end) - Panel
- Alex Tucker (Ginkgo Bioworks)
- Pamela Hines (Senior Editor at Science)
- Edward J. Hall (Professor of Philosophy at Harvard University)
- Tony Cijsouw (Neuroscience postdoc at Tufts University)
Scientific reproducibility
As Jeffrey Flier points out in his talk (1:24 - 29:31), there are a variety of ways of defining scientific reproducibility. For a study to be “reproducible” does not necessarily mean that someone has replicated it precisely. You can find a great discussion of different types of replication in Nosek and Errington 2017 and Schmmidt 2009, but for our purposes we’ll call a study reproducible if its findings can be predictably applied to future work.
Importantly, no one should expect that every study should be reproducible. Indeed, researchers are pushing the limits of knowledge as they develop new ways of understanding how the body works, generating new therapeutics, and sometimes formulating new biological concepts. However, rates of reproducibility for preclinical studies, as reported by drug companies attempting to apply the findings from these studies, are quite low (<30%, Prinz et al 2011, Begley and Ellis 2012). Recent efforts from the Reproducibility Project: Cancer Biology have produced similarly low rates of reproducibility. Failed attempts to apply irreproducible results to future work cost both time and money with estimates reaching in the billions of dollars (Freedman et al 2015). Finally, as Pamela Hines of AAAS points out on the panel, irreproducibility undermines public trust in science and research.
Nonetheless, I’d like to steer clear of saying we have a reproducibility “crisis” on our hands and instead provide a brief conceptual framework for the causes of irreproducibility with some solutions based on what I’ve learned. I hope you can apply some of these solutions to your own research and thereby help move the biological research enterprise towards greater reproducibility.
Cause 1: Inability to repeat previously performed experiments
Researchers should be able to follow one another’s protocols and use one another’s reagents with the confidence that they aren’t simply doing an experiment wrong. However, even simple procedures like pouring a plate or running a gel have intricacies that aren’t always mentioned in static written protocols. It is therefore a bit ridiculous to expect that one should be able to apply someone else's new and incredibly useful technique simply by following the abbreviated methods section found within a published manuscript.
Luckily, many organizations have been working to provide tools that make it easier to share in-depth information about reagents, protocols, analysis tools, and more. Being that this is the Addgene Blog, I’ll, of course, mention Addgene as a source of sequenced and quality controlled plasmids and viral vectors. When working with a plasmid from Addgene, you can be confident that you’re using the actual tool that you ordered and not a random aliquot pulled from a freezer.
As you’ll see in Steve Almo’s talk about work at the Institute for Protein Innovation (IPI) (49:45 - 1:18:45), there are also initiatives to create more reliable reagents for future research. I’ve talked to many researchers who have complaints about the unreliability of antibodies. IPI is working to address this issue directly by creating well characterised antibodies against all human extracellular proteins with community-based validation. They also have plans to expand to other protein technologies later.
Organizations such as BenchSci and FPbase are working hard to enable researchers by making it easier to find information about research tools. BenchSci collates information about antibodies while FPbase curates information about fluorescent proteins so you can best apply these technologies to your own research needs.
Other companies like NEB and ATCC can provide you with additional validated reagents, and, with these reagents in hand you’ve got a good foundation to begin experimenting. Protocols.io, JoVe, and Bio-protocol all work to provide more descriptive and interactive protocols that make it much easier to learn new experimental techniques. Electronic lab notebooks also play a role here by making it easier to retain searchable, thorough notes on how lab procedures were actually performed, potentially curtailing future confusion (and hours sifting through paper lab notebooks).
Once you’ve completed your experiment, you’ll need to analyze your results. As biological researchers deal with larger and larger datasets and use computer based analysis techniques, an important component of reproducibility is making sure that others can understand your analyses and repeat them. Companies like Code Ocean allow researchers to share code they’ve used for data analysis while other organizations like Dryad make it easier for researchers to share data thereby enabling you to do modified analyses on published data if you need to. Similarly, FigShare also makes it easier to share data through their curation of citable, linkable figures.
Cause #2: Hidden “negative” data
We all want to be able to tell a good story when presenting scientific findings. This makes sense on many different levels - it’s hard to process large amounts of information and, from a SciComm perspective, stories with links of causation from initial hypothesis through validation are easier to understand. The need for great stories has, however, made it difficult to publish so-called “negative” results that conflict with our original hypotheses. Looking for novelty in our stories, we are additionally averse to producing papers that repeat previously published results.
This over-reliance on the perfect story for a “high impact” publication can lead to the publication of rare flukes showing positive results when really there are no positive results to be found. Consider the hypothetical situation in Figure 1 where many scientists are performing similar experiments. Biology is messy enough that by chance, some small number of the scientists will get a result that confirms their hypotheses even if most other scientists do not. These scientists with positive results have done nothing wrong, but the problem is that only these positive results will be published. The publication record will then show that the tested hypothesis is correct (a great success story) even though the majority of the unpublished evidence shows otherwise. Others will go on to try to apply these findings in their own work ultimately wasting both time and money.
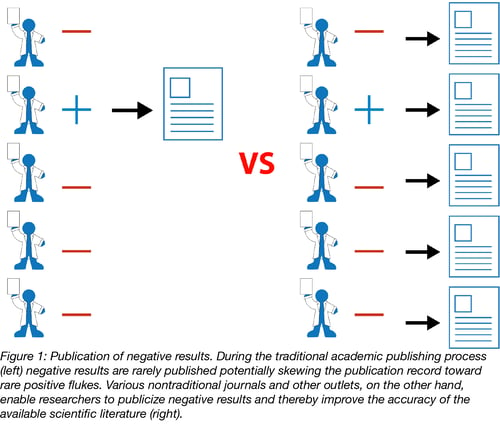
The obvious solution to this problem would be for traditional journals to publish negative data, but this is a big ask and, from the journal’s perspective, probably not what most readers want to read.
As Susanna Bachle points out in her talk (29:31 - 49:45), there are many ways to publicise negative data - we don’t have to rely on traditional journals. There’s social media - science Twitter is very active with many scientists sharing their unpublished data. There are blogs - both hosted by individuals and by organizations like Addgene (a good example is our series of blog posts on the defunct genome editing technology NgAgo). In addition, there are organizations like ReFigure which aims to consolidate published and unpublished experiments on similar subjects in a single, searchable space. Finally there are new publishing outlets like F1000, PeerJ, PLoS, and bioRxiv that make it easier to publish (or pre-publish) sound data even if it isn’t considered immediately impactful.
Down the line we may see further adoption of policies that promote reproducible publishing practices. One example is the publication of peer reviews so we can see how and why papers change throughout the review process. This will hopefully push reviewers towards more productive criticisms that focus less on perceived novelty and impact. Journals could also begin requiring reproducibility checklists that at least make it obvious that reproducibility is a priority for the journals themselves. Within universities, we’ll also hopefully see a shift toward hiring and promotion practices that focus on reproducibility and ensuring the health of the scientific enterprise and not just impact.
Cause #3: Poor experimental design
It’s easy to get excited about positive results. Not only is it scientifically and intellectually affirming to have a hypothesis confirmed, it also means that you’re more likely to get a publication (for better or worse, the currency for success in academic research). Initial positive results and the excitement they generate can, however, bias us. We’ve all heard stories and read papers wherein a researcher tries 5 separate experiments at once to get confirmatory data for a hypothesis and presents only the results that align with the original hypothesis. Other practices that fall into this category include performing experiments with too few replicates leading to suspect statistical significance (you can read more about this problem in Button et al 2013 and Ioannidis 2005) and changing an experimental set-up partway through an experiment in an attempt to get results that align better with an initial hypothesis.
To be clear, I don’t think that the majority of researchers do these things with nefarious goals in mind. Indeed, it’s not always obvious that we’re doing things that make it more likely that we’ll find a fluke when we’re simply trying to get our experiments to work, but part of good experimental design is recognizing that we’re biased and taking steps to prevent our biases and goals from effecting our experiments.
How do we engender better experimental design? The summation of many things I’ve read comes to this: we need others to review our experimental designs and we need to be transparent about the reasons we alter these designs if we do so.
The most extreme version of this (and possibly the most effective if it were more practical) is pre-registration and review of experimental design as part of the publication process (Nosek et al 2018). In this ideal situation, a researcher would submit an experimental design to a journal prior to performing the experiment (or set of experiments), the design would be reviewed, and, if approved, the researcher would get in-principle acceptance of the final manuscript regardless of whether the ultimate results were positive or negative.
I can feel you rolling your eyes, but even if this ideal form of pre-registration doesn’t come to fruition any time soon, the Center for Open Science does host a platform for pre-registration wherein researchers can submit designs and transparently reveal them upon completing an experiment. There’s not necessarily in-principle acceptance, but readers of pre-registered publications can have more confidence that the experiments they’re reading about weren’t changed halfway through in order to chase a positive result.
Beyond these more formal systems, individual labs can certainly institute better experimental design practices including talking to statisticians when necessary and encouraging interlab peer review of experimental design. These are simple steps but could go a long way to encouraging researchers to take a second and think about how their biases affect their experimental designs.
There are many more facets to reproducibility issues in the biological sciences. I’d highly recommend listening to the panel discussion to learn a bit more about how the publishing community views reproducibility, how industry relies on reproducibility, and how you can push for more reproducibility in your own lab. While reproducibility issues are complicated, we have many potential solutions that could save both your lab and the entire research enterprise time and funds. It’s time to start putting them into practice!
Additional Resources
- SciLine at AAAS - A service connecting journalists and scientific experts discussed by Pamela Hines
- Minisymposium on Reproducibility Booklet - Including descriptions of many organizations promoting reproducibility
- List of Tools for Reproducibility at Protocols.io
Additional Resources on the Addgene Blog
Topics: Scientific Sharing, Reproducibility






Leave a Comment