%20576.png?width=350&name=SnapGene%20Logo%20(White)%20576.png) This post was contribued by guest bloggers Aline and Benjamin Glick from SnapGene.
This post was contribued by guest bloggers Aline and Benjamin Glick from SnapGene.
SnapGene was created to meet a need. While there were software tools available to biomedical researchers manipulating DNA sequences on a daily basis, many found these tools inadequate for planning, visualizing, and documenting their procedures. Preventable errors in the design of cloning strategies set experiments back days or even weeks. Primer design was done painstakingly by hand. Records of plasmid construction were often incomplete or nonexistent. In the 21st century, many molecular biologists didn’t know the complete sequences or properties of the DNA molecules they were using.
SnapGene was created to alleviate these problems through good software design. But what makes software good? Fortunately, that question has been thoroughly answered by experts in human-computer interaction (HCI), and we have adhered rigorously to HCI principles. For every task, we envision what the user wants to do and make the path to accomplishing their goals as intuitive and painless as possible. Instead of crowding the interface with every possible option, we place the most important controls front and center, and make specialized controls available when needed. SnapGene has been engineered to be easy and enjoyable to use. Development of software with these qualities is an ongoing process that involves iterative refinements in response to customer feedback.
An example of this approach is SnapGene’s algorithm for detecting common features. This algorithm enables one of SnapGene’s most popular aspects - its ability to annotate a raw plasmid sequence and display frequently used genes and control elements. Development of this tool required creating a database of common features, and devising rules for identifying a feature even when the match is imperfect.
SnapGene's feature database
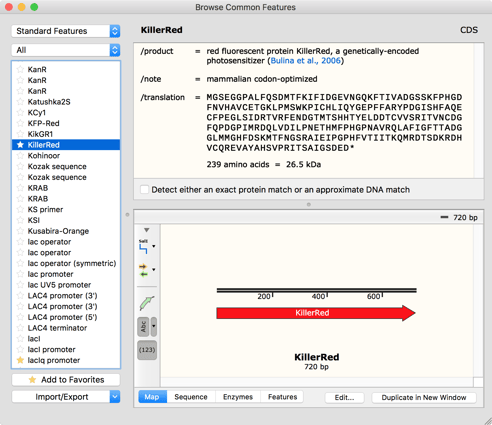
The source of common features was our collection of popular plasmid sequences. These plasmids contain features such as antibiotic resistance markers and replication origins, but there is extensive heterogeneity in the feature sequences due to genetic drift and the use of genes from different microbial strains. It proved to be impractical to catalog every variant of a feature. Instead, we identified common variants, and then crafted a detection algorithm that tolerates occasional mismatches or indels. Empirical tests indicated that a reasonable rule is to require at least 96% sequence identity when detecting a reference feature. For a coding sequence feature that may be used to make fusion genes, detection needs to occur even if one or two codons are missing at the beginning or end of the feature. With the increasing popularity of gene synthesis, many researchers now use codon-optimized versions of common coding sequence features, so our detection system was enhanced to allow searches for a perfect protein sequence match even when the DNA sequence has changed.
Identifying plasmid control elements
Coding sequence features are relatively straightforward to define, but for control elements such as promoters and transcription terminators, the boundaries are less obvious. We found that the annotated boundaries for control elements in commercial plasmids were inconsistent and sometimes clearly wrong. In an effort to be rigorous, we dug into the original literature, some of it decades old, to provide reliable feature annotations.
Even with these extensive efforts, such an algorithm has limitations. For instance, it could miss a common feature if the sequence differences exceed the threshold. This issue is addressed by adding more variants to the database. Another limitation is that by tolerating mismatches, our algorithm could annotate a feature inaccurately. The best example is fluorescent proteins, which often come in closely related versions that have different properties. To prevent misidentification, we augment the database with new fluorescent protein variants.
The future of DNA feature identification
What’s next for common features? We plan to continue updating the feature database. Powering the plasmid maps on Addgene.org with SnapGene Server is useful because it helps us find which common features need to be added to the database – we will work to fill any common annotation gaps in the plasmids available through the Addgene website. In addition, two initiatives are in the pipeline. First, now that SnapGene supports “Collections”, which are shareable databases of DNA and protein sequences, the next step is to enhance Collections so that a group or organization can annotate their sequences from a shared database of custom common features. Second, customers have asked for a way to annotate the genomes of newly sequenced bacterial or viral strains. We are working on a way for SnapGene users to apply features from a reference strain to newly sequenced strains through our mismatch tolerant detection algorithm and thereby speed up the annotation process.
Many of the best ideas come from engaging with users of the software. We welcome suggestions about common features from anyone who uses SnapGene or the free SnapGene Viewer companion product. If you don’t use SnapGene yet, you can download a free trial. If you notice that a feature is missing from our database, or if you see an opportunity to harness common feature detection in a new way, please contact us at snapgene.com.
Many thanks to our guest bloggers Aline and Benjamin Glick from SnapGene!
 Ben Glick is President and Chief Scientist at SnapGene, and also a Professor of Molecular Genetics and Cell Biology at the University of Chicago. He was an early depositor at Addgene, and conceived SnapGene because he needed it for his own lab.
Ben Glick is President and Chief Scientist at SnapGene, and also a Professor of Molecular Genetics and Cell Biology at the University of Chicago. He was an early depositor at Addgene, and conceived SnapGene because he needed it for his own lab.
 Aline Glick is VP of Product Management at SnapGene. She is passionate about building software that people love to use, and likes to start at the beginning, shepherding a new product from the idea stage through all of its post-launch iterations.
Aline Glick is VP of Product Management at SnapGene. She is passionate about building software that people love to use, and likes to start at the beginning, shepherding a new product from the idea stage through all of its post-launch iterations.
Additional Resources on the Addgene Blog
- Read about our SnapGene powered plasmid maps
- Learn how we use SnapGene during the plasmid quality control process
- Check out full plasmid sequencing at Addgene
Additional Resources on Addgene.org
Topics: Plasmid Elements, Other Plasmid Tools, Plasmids







Leave a Comment