This blog post was contributed by guest blogger Kate Palozola
Traditional lab notebooks just won't cut it for bioinformatics. All kinds of biologists are finding themselves using computational approaches to analyze large data sets (myself included) and we are faced with finding the best system to document these types of analyses and their results. We are adept at recording wet-lab experiments using a “traditional” lab notebook; however, keeping track of computation work comes with new sets of challenges. One challenge with computational analyses is to keep track of why you are doing what you are doing. Another common challenge is to keep track of what works, and what does not work. Careful documentation will keep you on task and will prevent you from getting lost in the wide word of informatics.
I am a molecular biologist who began coding for my thesis project a few years ago when my advisor and I decided to do a massive RNA-seq analysis. I had never interacted with a computer at the command line, and so was really starting from the beginning. Luckily, a fellow graduate student and a data analyst in our lab took it upon themselves to teach me Python, a programming language favored by biologists. Almost as soon as I began, I realized that I needed a system to keep track of the files that I was continuously generating.
I quickly found that there is no one way to keep a virtual lab notebook for bioinformatics. In fact, there are endless ways and everyone finds their own. Here, I simply outline what works for me, and I hope that it is helpful to those of you just getting started. The practices suggested here may seem tedious at first, especially since you’ll want to dig in to the computation, but they will serve you well as you perform analysis after analysis after analysis…
Treat every analysis like a wet lab experiment
I use the text editor TextWrangler to record the following, which are later printed for my physical lab notebook, along with any figures generated from the analyses:
- Goal: Before you begin, briefly state the goal of the analysis, including background on other analyses that have inspired you to perform this analysis. Having a clearly stated, specific goal for each analysis will help you locate relevant information in the future.
- Approach: Outline a brief overview of the approach that you will take to help you plan your analysis. There is no need to go into a lot of detail – the details will be in the scripts that you run. Rather, this is where you outline your logic and the scripts and input files that will be used to perform each task.
- Conclusion: Always, always, write a brief conclusion of your analysis, even if your conclusion is “this approach is not ideal because…” Including a conclusion for each analysis or task will keep you from repeating your work or making similar mistakes in the future.
Stay organized
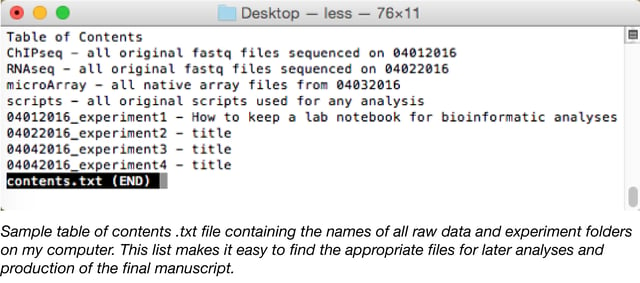
- Set up a table of contents in the top directory: A directory is a file system in the computer and is just another name for a folder. For instance, your Desktop directory contains the folders and files that you see on your desktop. The location of your master directory or folder for all of your analyses is up to you, but should contain a table of contents that lays out which experiments can be found in each folder. This can be a simple .txt document. Record the name, date, and location of all analyses (see above).
- Give every experiment its own directory: In addition to output files, this is also where you can store any power point, excel, or other files relevant to the project.
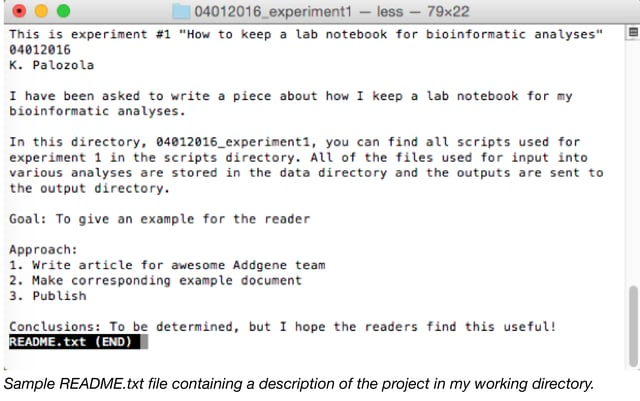
- README: The README.txt document is invaluable. In the top directory of every experiment, immediately write a text document with a brief description of the directory contents, at a minimum. This may or may not be where you also record your goal, approach, and conclusions (see above).
- Keep original files in a separate folder: You will often use the same data files for multiple analyses. Rather than copying these files to your working directory (the folder that you are currently working in) every time you use them, leave them in their own folder. Doing so will ensure that you are in fact using the same data for all of your analyses. The same is true for scripts.
Naming conventions
- Give every experiment a number: Numbers are a short, easy way to name files so that you know which files go together. I also prefer the number system to using dates as labels, since I often work on multiple projects in a given day. For example, instead of naming a file “output” name the file “1_output” so that you know that the file is the output of the analysis performed in experiment 1.
- Use camelCase: In camelCase, each subsequent word in a file name after the first word begins with a capital letter and words are not separated by spaces. File names with more than one word should be named in the camelCaseFormat since spaces between each word can make it difficult to accurately call a file.
- Version control your commonly used scripts: If you edit a general purpose script for a single use, save the script as originalName_descriptionOfEdit in the same directory where it was run. This technique leaves a trail that further helps you keep track of your exact changes. Alternatively, you can simply make a note in your methods section about the edit that was temporarily used. Just be sure that the original code remains in tact!
Take notes
- Comment in your scripts: Comments on what a given line of code does are very useful for those that are just starting out, and helps clarify the role of the code.
- # This is a comment in python, R, perl, and ruby
- // This is a comment in C++ and Java
- Command line history: Tracking command line history by recording all commands entered is useful for beginners who are still learning the basics. However, these notes take up a lot of space and don’t tell you why you did something or what the result was, so try not to do this.
These are the simple rules that I try to follow in my own research. Again, some of this may not apply or may not work for you. The best way to find a system that does is through trial and error, and by asking for tips from others. Comment below to add your tips!
Many thanks to our guest blogger Kate Palozola!
 Kate Palozola is currently a graduate student at University of Pennsylvania. She is particularly interested in science communication and epigenetics. Follow her on twitter @kc_palozola.
Kate Palozola is currently a graduate student at University of Pennsylvania. She is particularly interested in science communication and epigenetics. Follow her on twitter @kc_palozola.
Additional Resources on the Addgene Blog
- Read Our Tips to Keep Your Lab Notebook Organized
- Learn about Software You Can Use to Save Time and Track Your Experiments
- Try out the CRISPR Software Matchmaker
Additional Resources on Addgene.org
Topics: Other, Miscellaneous






Leave a Comment