 Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. Below you'll find our hot plasmid articles from April 2018. If you'd ever like to write about a recent plasmid deposit please sign up here.
Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. Below you'll find our hot plasmid articles from April 2018. If you'd ever like to write about a recent plasmid deposit please sign up here.
No time to read? Listen to our Hot Plasmids Podcast!
TRIM-Away: Targeted endogenous protein degradation
Article contributed by Mary GearingListen to the TRIM-Away podcast segment
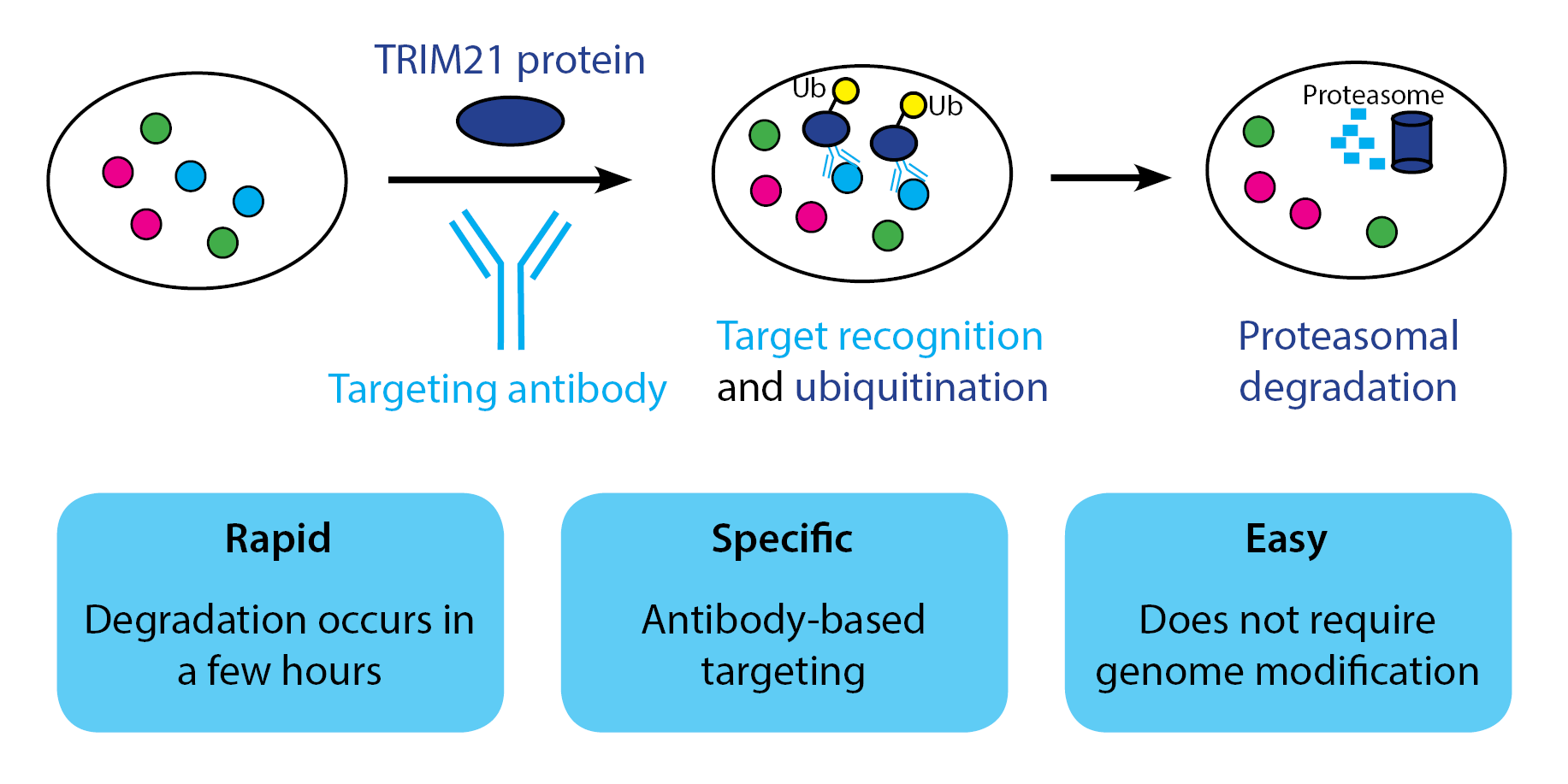
CRISPR and RNAi have helped researchers alter DNA sequence and RNA expression - but what if you just want to change protein levels? Until recently, no such tool existed for rapid, customizable protein degradation. Enter TRIM-Away - an antibody-based system developed by the James and Schuh labs. Clift et al. applied TRIM-away to nine proteins in ten ddifferent cell types, including difficult-to-manipulate primary cells, achieving knockdown within 1-2 hr of reagent delivery. TRIM-Away is suitable for both short-and long-lived proteins, and it can even distinguish between protein variants when used with a variant-specific antibody.
TRIM21, an E3 ubiquitin ligase, is the basis of TRIM-away. In normal physiology, TRIM21 recruits ubiquitin and the proteasome to destroy antibody-bound pathogens. Clift et al. found that addition of exogenous TRIM21 protein along with an antibody targeting the protein of interest led to rapid, targeted degradation that is dependent on proteasome function. TRIM-away compared favorably to drug inhibition, with TRIM-away targeting of mTOR producing similar results to rapamycin inhibition. To assess TRIM21 toxicity, Clift et al. created stable cell lines overexpressing TRIM21, finding that TRIM21 overexpression did not affect cell proliferation. These cells also did not contain changes in expression of TRIM21-associated genes, indicating that TRIM21 overexpression does not aberrantly activate the proteasome.
Clift et al. tested multiple methods for TRIM-Away delivery. For small cell populations, co-microinjection of TRIM21 protein and the target antibody works well. In larger cell populations, you can deliver these components together using electroporation. In these bulk cell experiments, Clift et al. observed no stress or damage upon TRIM-Away, and the electroporated cells re-adhered to the culture plate within 4 hr of treatment. In some primary cell lines, endogenous TRIM21 is sufficient for TRIM-Away, and you only need to deliver the target antibody. TRIM-Away reagents and reporters are available from Addgene - we hope they will TRIM down the difficulty of your protein knockdown experiments!
The TRIM-Away plasmids can be found here.
- Clift et al. Cell. 2017. PubMed PMID: 29153837.
Endosomal pH sensors for STED nanoscopy
Article contributed by Eric J. Perkins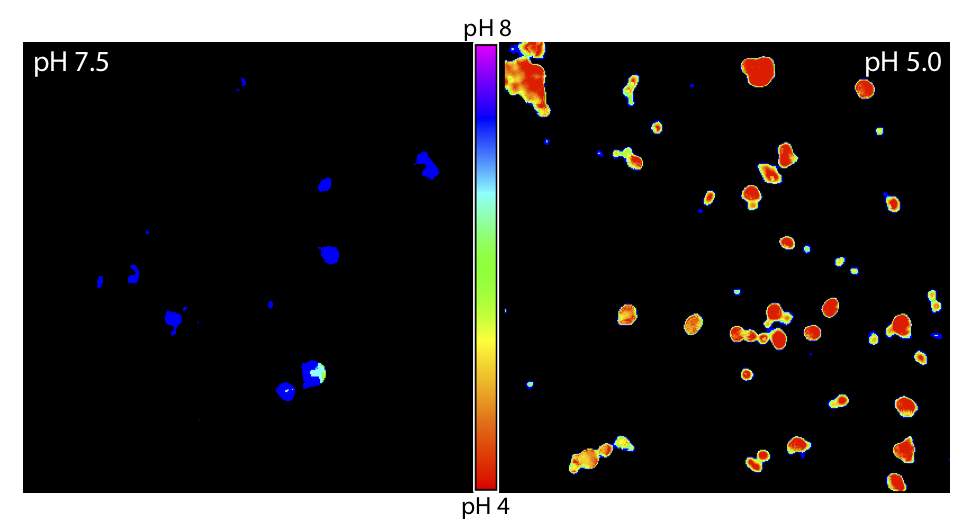
Listen to Endosomal pH Sensors podcast segment
The first Addgene plasmid deposit from Professor Stefan Hell, who won the 2014 Nobel Prize in Chemistry for his contributions to super-resolution light microscopy, comes from his recent Nature Communications article “SRpHi ratiometric pH biosensors for super-resolution microscopy.” Endosomes are historically difficult to study in living cells using conventional light microscopy due to their small size (<250 nm), and studying endocytosis is further complicated by the reliance on pH-based biosensors used to detect the maturation of neutral vesicles to more acidic lysosomes. Though a number of biosensors existed that met some of the criteria necessary for studying endosomes, the Hell lab designed their super-resolution pH indicators (SRpHi) biosensors to meet all of them. These probes had to target endosomes, be exceptionally bright (which is necessary for effective STED), and be photostable over a broad pH range. The four biometric probes successfully targeted endosomes under a range of pH conditions in HeLa and MEF cells.
For further reading on fluorescent microscopy techniques, the first author of this article, Dr. Douglas Richardson, recently contributed a relevant post to the Addgene blog.
Find the SRpHi Biosensors here.
- Richardson et al. Nat Communications. 2017. PubMed PMID: 28924139.
FIRE-Cas9 for inducible interrogation of the epigenome
Article contributed by Joanne KamensListen to the Fire-Cas9 Podcast Segment
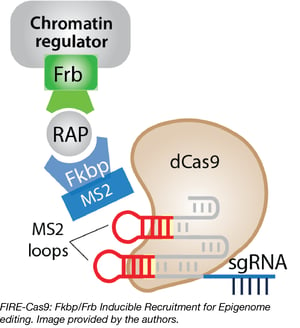
Jerry Crabtree’s lab has developed a new CRISPR/Cas9-based system to study epigenetic modulation. While catalytically inactive cas9 (e.g. dCas9) has been fused to a variety of protein modules to repress or activate gene expression, sometimes incorporating chemically inducible approaches to provide temporal control, this new system, Fkbp/Frb Inducible Recruitment for Epigenome editing by Cas9 (FIRE–Cas9), combines these approaches to enable rapid and reversible recruitment of chromatin complexes to any genomic locus in any cell type. By varying which sgRNAs are used with FIRE-Cas9, experiments can be designed to target multiple loci at once. In addition, the approach allows minute-by-minute analysis of the kinetics of chromatin regulation and subsequent mathematical modeling. Its reversibility allows precise and quantitative analysis of epigenetic memory at nearly any locus.
The FIRE-Cas9 plasmids can be found here.
- Braun et al. Nat Communications. 2017. PubMed PMID: 28916764.
Plasmids for independent control of gene expression in yeast
Article contributed by Tyler J. FordListen to the Synthetic Yeast Transcription Factors Podcast Segment
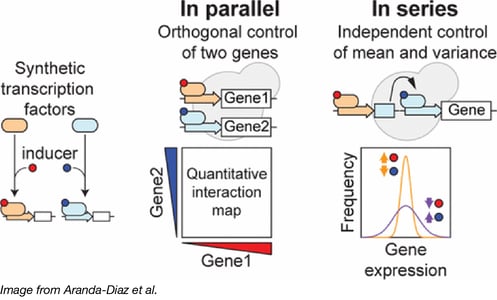
Researchers from the El-Samad lab recently deposited plasmids encoding synthetic yeast transcription factors that can independently control expression of your genes of interest. These transcription factors consist of the GAL4 or ZIF268 DNA binding domains fused to either the hER or hPR ligand binding domains and the MSN2 transactivation domain. Transcription factor activation via the appropriate ligand - estradiol for hER or progesterone for hPR - leads to nuclear localization, binding to the appropriate promoter sequence and activation of gene expression. In addition to plasmids encoding these synthetic transcription factors, the El-Samad lab has deposited plasmids encoding fluorescent reporters and other proteins controlled by GAL4 and ZIF268.
To demonstrate independent control of gene expression, Aranda-Díaz et al transformed yeast with both a GAL4/estradiol controlled transcription factor and a ZIF268/progesterone controlled transcription factor as well as reporters specific to each transcription factor. They then induced cultures of these transformants with various concentrations of estradiol and progesterone and showed that reporter expression is dependent only upon its cognate inducer. Since induction via estradiol and progesterone do not affect each other, these systems can be used together.
Aranda-Diaz et al additionally show that it is possible to use synthetic transcription factors to control the level of random variation, or noise, in reporter expression. To do so, one places reporter expression under the control of two separate synthetic transcription factors in series and independently changes inducer levels. That is, if transcription factor 1 controls expression of transcription factor 2, which directly controls expression of a reporter protein, it is possible to change the amount of noise in reporter gene expression by modulating the induction of transcription factors 1 and 2. See Aranda-Díaz et al. for more information. The El-Samad lab predicts that this level of control will enable future researchers to better simulate physiological gene expression regimes that cannot be easily achieved with classical gene overexpression and knockout techniques.
Find the plasmids from Aranda-Díaz et al.
- Aranda-Díaz, et al. ACS Synth Biol. 2017. Pubmed PMID: 27930885.
Topics: Other Plasmid Tools, Plasmids






Leave a Comment