Every few months we highlight a subset of the new plasmids and viral preps in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.

Here's what you'll find in this post:
- Improving flexibility for in vivo proximity labeling
- Increased neuronal expression using Cre-mediated AAV Targeted Evolution method (M-CREATE)
- dCas9-SSAP with reduced error rate for long sequence knock-ins
- Visual barcodes for live cell clonal multiplexing
Improving flexibility for in vivo proximity labeling
By Ashley Waldron
Proximity-dependent labeling has become a powerful approach for proteomic mapping in both cell culture and whole organisms (see our 2019 overview). Until recently, the approach has been hampered by the need to tag each protein of interest and, for in vivo work, generate a dedicated transgenic line. Using zebrafish as a model organism, the Parton Lab recently created a flexible system that makes use of pre-existing transgenic lines, termed BLITZ (Biotin Labeling In Transgenic Zebrafish) (Xiong, et al., 2021).
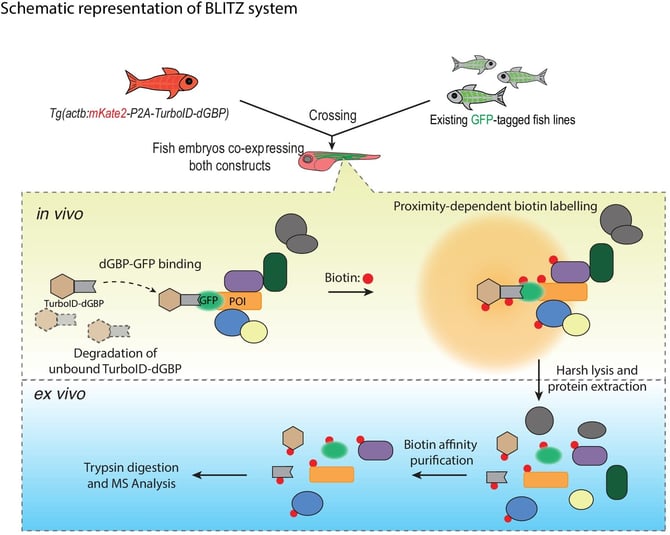 |
| Fig. 1: Schematic showing the workflow for the BLITZ system. Image from Xiong et al. 2021. |
BLITZ uses the biotin ligase TurboID fused to a conditionally stabilized GFP-nanobody rather than a specific protein of interest. This fusion protein can then be co-expressed in essentially any transgenic zebrafish line expressing a GFP-tagged version of a protein of interest. The binding of the GFP nanobody to its target stabilizes the TurboID-nanobody protein and allows TurboID to start labeling proteins in close proximity to the GFP-tagged protein (albeit slightly offset due to the nanobody-GFP interaction). The modularity of the system provides researchers better flexibility and makes the approach more accessible, as it reduces the need to generate “single-use” transgenic lines. Instead, researchers can generate and maintain transgenic lines that can be used for other experiments. And, while the BLITZ system was designed and optimized with zebrafish in mind, it can easily be applied to other systems.
Xiong, et al. eLife. 2021. https://doi.org/10.7554/eLife.64631
Goertsen D, et al. Nature Neuroscience. 2022. https://doi.org/10.1038/s41593-021-00969-4.
Increased neuronal expression using Cre-mediated AAV Targeted Evolution method (M-CREATE)
by: Brian O'Neill
The Viviana Gradinaru Lab has previously developed the multiplexed version of the Cre-mediated AAV Targeted Evolution method (M-CREATE), in which one applies both positive and negative selective pressures for novel capsids. In a continuation of that work, they have found at least two variants that display increased neuronal expression in the brain while having decreased expression in the liver after systemic delivery. Importantly, this is true in both mice and nonhuman primates (for AAV.CAP-B10, for example). Given that the liver is an immunologically active organ, this advance is likely to afford the ability to deliver more therapeutics to the CNS, and with reduced systemic side effects, by allowing the user to select for variants that infect the CNS while selecting against those that also infect the liver.
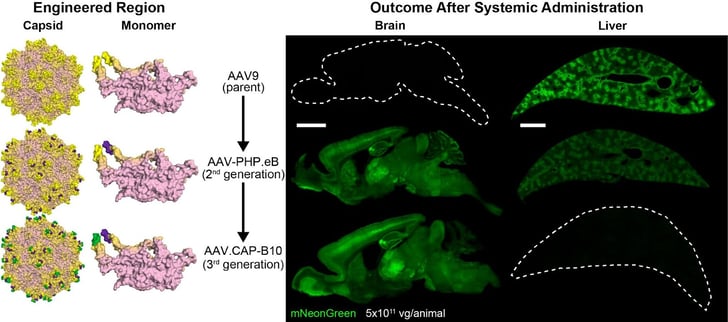 |
| Fig. 2: Summary of the generational evolution of the AAV capsids (left) and transduction patterns (right). Both AAV-PHP.eB and the AAV.CAP-B capsids are derived from AAV9. Of those shown here, only AAV.CAP-B10 causes brain wide expression, with no liver expression, after retro-orbital sinus vein injection of a CAG-driven mNeonGreen construct in mouse. Image from Goertsen D, et al., 2022. |
Goertsen D, et al. Nature Neuroscience. 2022. https://doi.org/10.1038/s41593-021-00969-4.
dCas9-SSAP with reduced error rate for long sequence knock-ins
by: Lucie Wilson
Le Cong’s lab has developed a method using dCas9-SSAP to knock-in long sequences with reduced risks of unwanted mutations in mammalian cells. Long sequences traditionally have higher risks of off-target mutations. These plasmids contain catalytically inactive dCas9 and microbial SSAPs (dCas9-SSAP) which insert genes in a cleavage free manner. The dCas9-SSAP system reports an efficiency of up to 20%, comparable to commonly used CRISPR/Cas9 methods for large inserts. However, this system has a much lower error rate. For a one kilobase insertion, the researchers reported an editing error rate of less than 0.3%, both on and off target. Across all loci tested, error rates overall ranged from 0.3% to 10%, in comparison to the 62-90% error rate seen with Cas9 editors. The lab believes that this system could potentially be packaged into an AAV system. dCas9-SSAP will make the editing of kilobase-scaled sequences more efficient and less risky.
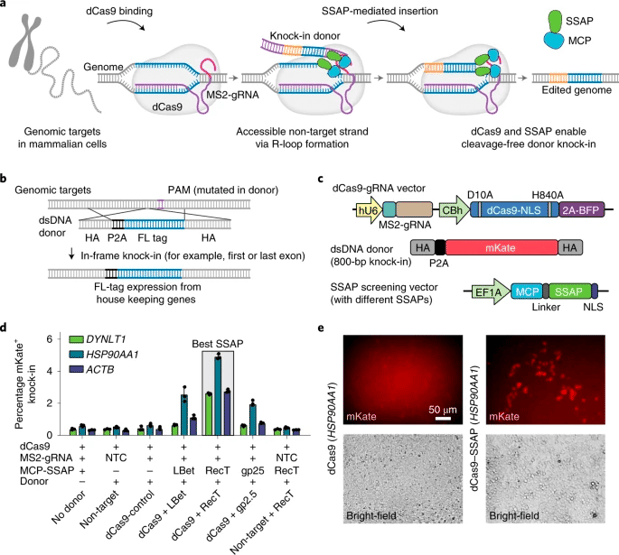 |
| Fig. 3: Development of a cleavage-free dCas9-based gene editor using microbial SSAPs. Image from Wang, et al, 2022. |
Find dCas9-SSAP plasmids here!
Wang et al. Nat Cell Biol. 2022. https://doi.org/10.1038/s41556-021-00836-1
Visual barcodes for live cell clonal multiplexing
by: Rachel Leeson
Multiplexed images in live cells remains a challenge due to multiple factors, including limitations of number of flourescent proteins that can be inserted into a cell without disrupting its functions. Recently, Ravid Straussman’s lab has developed visual barcodes for clonal multiplexing of cells in live imaging application. Each visual barcode consists of a fluorescent protein coupled to a specific subcellular localization peptide. By combining three different localization peptides with four different fluorescent proteins, and inserting each visual barcode into its own subclone, they were able to develop a 12-plex multiplex imaging system. Mixing the subclones within a single well reduced well-to-well variability, while the readout was able to be deconvoluted using freeware.
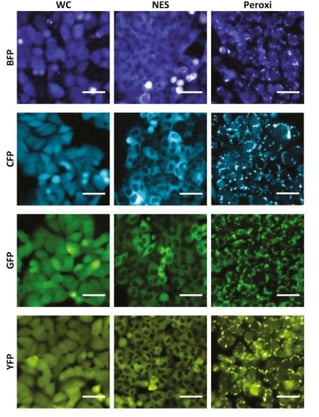 |
| Fig. 4: Images of 12 visual barcodes in use in A375 cell line. Image from Kaufman, et al, 2022. |
This new barcoding technology would allow researchers to avoid the off-target effects of hi-multiplexing within in a cell, while potentially allowing for up to 20-plex experiments with one visual barcode per cell and up to 200-plex with two barcodes per cell. The researchers validated their work with the “signalome,” a 12-plex mixed-subclone system using mStrawberry, localization peptides, and reporters that drive the expression of the fluorescent protein by a specific transcription response element (TRE) or kinase translocation reporters (KTRs).
Find visual barcode plasmids here!
Kaufman, et al. 2022. Nature Comm.https://doi.org/10.1038/s41467-022-30008-0
Topics: Hot Plasmids






Leave a Comment