 Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
AAV delivery of split base editors
Article contributed by Jennifer Tsang
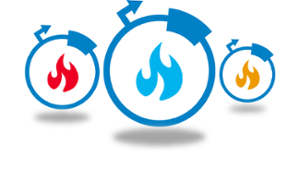
AAVs are a great gene therapy delivery mechanism, but their small packaging capacity of 4-5 kb limits the delivery of larger transgenes, such as base editors which have a gene size of ~5.2 kb. To overcome this problem, David Liu’s lab developed split cytosine and adenine base editors that can be reconstituted in vivo to form a protein identical in sequence to the unmodified enzyme. Each half of the base editor is fused to a fast-splicing split intein and co-infected into the cell. Intein splitting removes any exogenous sequences and regenerates a native peptide bond at the split site.
 |
| Figure 1: The base editor is split into two plasmids and co-transfected into cells for reconstitution. Image from Levy et al., 2020 with permission. |
Though the lab suspected that the co-transduction by two viruses would lower editing efficiency, the split editors transduced at levels similar to that of a single AAV expressing GFP. The lab tested the base editors in vivo and achieved therapeutically relevant editing efficiencies in the mouse brain, liver, retina, heart, and skeletal muscle. The dosing required to achieve these efficiencies was also between 1013 - 1014 vg/kg, a dosage comparable to those currently used in human gene therapy trials. They further tested the base editors by correcting a mutation that causes Niemann-Pick disease type C, directly demonstrating the therapeutic potential for the split base editors.
Find the split base editors at Addgene.
Levy et al., Nature Biomedical Engineering, 2020. PubMed PMID: 31937940.
Detection of neural activity using a red fluorescent genetically encoded voltage indicator
Article contributed by Shreya Vedantam
Genetically encoded voltage indicators (GEVIs) are proteins that detect and output the voltage changes in neural activity when paired with appropriate fluorescent optics. Existing red fluorescent voltage indicators, however, fall short compared to GFP-based sensors in terms of response and brightness. They also do not spectrally separate from green sensors and blue-light-activated optogenetic actuators.
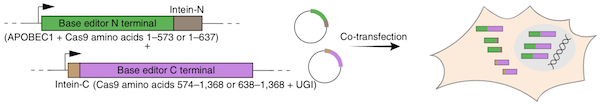
To overcome this overlap, Yiyang Gong’s lab created two plasmids containing a red-shifted Ace-mScarlet, a fusion of a voltage-sensitive inhibitory rhodopsin (Ace2N) with the red fluorescent protein (mScarlet) with either a seven or eight amino acid linker between them. The Ace-mScarlet voltage indicator works like many FRET-opsin GEVIs. With an increase in the neuron’s membrane potential, Ace absorbance increases and results in higher quenching of mScarlet emission.
 |
|
Figure 2: Ace-mScarlet is a fusion between the voltage-sensitive inhibitory rhodopsin Ace2N and mScarlet. Ace2N quenches a proportion of mScarlet's emission via FRET. Image from Beck and Gong, 2019. |
Broadening the spectral diversity of GEVIs can enable resolving single action potentials and further studies of optical neural circuit manipulations when combined with existing green-fluorescent GEVIs and blue-light-sensitive channelrhodopsins.
Find the Ace-mScarlet voltage indicator plasmids at Addgene!
Beck et al., Sci Report 2019. PubMed PMID: 31685893.
A MoClo toolkit for cyanobacteria
Article contributed by Andrew Hempstead
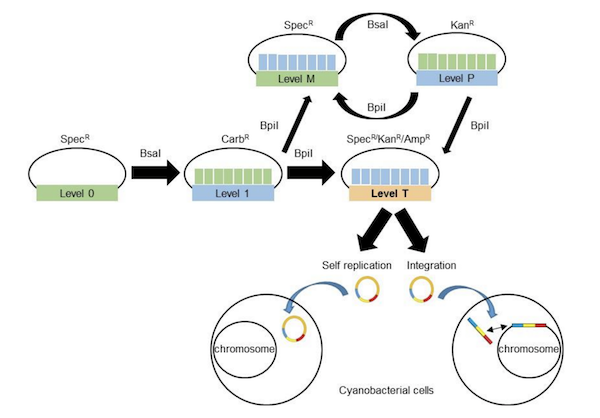
The Modular Cloning (MoClo) system allows synthetic biologists to use Type IIS restriction sites to easily assemble multiple DNA fragments. This system has been used to construct plasmids for use in model organisms such as E. coli and yeast. Because this approach uses standardized parts, it allows for greater reproducibility as scientists can assemble parts from different labs and experimental systems in their own research.
Cyanobacteria, a phylum of photosynthetic bacteria, has increasingly been recognized for its biotechnological applications including conversion of CO2 into water and electrical energy generation. Alistair McCormick’s lab recently developed the CyanoGate Kit for use of the MoClo system in cyanobacteria. This kit contains 96 parts including terminators and promoters, CRISPRi parts, selectable markers, transformation acceptor vectors, and sequences for genomic integration by homology directed repair. Using parts from this kit, the McCormick lab generated knock-out and knock-in mutants, determined protein expression levels from different promoters, and inhibited gene expression using CRISPRi. The kit functions in the model cyanobacterium Synechocystis sp. PCC6803, and the fast growing strain Synchococcus elongatus UTEX2973, showing the potential for broad use. This kit is designed for use with acceptor vectors from the MoClo Toolkit from Sylvestre Marillonnet’s lab to develop synthetic biology tools for cyanobacteria.
|
|
| Figure 3: Assembly of plasmids using the Cyanogate Kit and subsequent transformation into cyanobacteria. Image from Vasudevan et al., 2018. |
Find the CyanoGate kit at Addgene.
Vasudevan et al., Plant Physiol 2018. PubMed PMID: 30819783.
Light up CreLite: a red-light inducible optogenetic tool for precision in cell lineage tracing
Article contributed by Erin Sanders
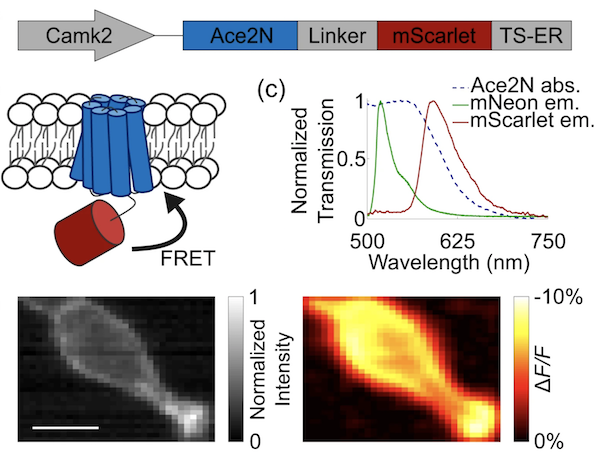
CreLite, designed by the Eisenhoffer lab, is a new tool that increases temporal and spatial precision of gene expression and reduces toxicity as compared to previous Cre/lox or drug-induced Cre systems (ex: tamoxifen). As the name suggests, CreLite combines the Cre/lox system with optogenetics. Cre activity is controlled by red light by splitting Cre and fusing each half with PhyB or PIF6, phytochrome-like fusion proteins from the plant Arabidopsis thaliana. In the presence of red light (650 nm) and its prosthetic group PCB, PhyB and PIF6 dimerize, bringing each terminus of Cre together to restore Cre activity. As soon as red light or PCB is no longer present, the fusion proteins separate, inactivating Cre. This is critical for reducing Cre toxicity in long-term experiments.
 |
| Figure 4: The C-terminus of Cre (CreC) and the N-terminus of Cre (CreN) are fused to the truncated Phytochrome B (PhyBΔ) and the active protein binding domain of Phytochrome B interacting factor 6 (PIF6APB), respectively. The cofactor analog, phycocyanobilin (PCB), is required for PhyBΔ to be functional. Exposure to red light induces PhyBΔ and PIF6APB binding, bringing the two halves of Cre together. Image from Eisenhoffer et al., 2019. |
This novel tool is practical in cell and organ culture, as well as cell lineage studies. Eisenhoffer and his colleagues evaluated CreLite for cell tracing using the transgenic zebrafish model, ubi:zebrabow. In this system, cells express various fluorescent proteins controlled by Cre recombination. Each cell can have multiple copies of the transgene that recombine independently and results in a variety of colors depending on the combination of fluorescent proteins expressed. CreLite-mediated recombination was found in both deep and superficial tissues of zebrafish studied, such as the brain, eye, muscle and epithelial cells, as well as in different stages of development, ensuring the breadth and versatility of this tool.
Yen et al., bioRxiv 2019. DOI: 10.1101/823971.
Topics: Hot Plasmids, Other Plasmid Tools, Plasmids






Leave a Comment