If you’ve ever used Golden Gate Assembly for cloning, you might be familiar with the rules of thumb for designing your overhang sets. But are those rules the best way to design GGA overhang sets, particularly for high-complexity reactions?
The researchers over at New England Biolabs® thought otherwise! They suspected a data-driven approach might allow for an increase in the complexity. In 2018, they published a paper profiling four base overhang ligation (Potapov et al., 2018), which showed that the five traditional rules didn’t need to be strictly followed in order to generate high-fidelity reactions. (Need a refresher? The five rules are: (1) don’t use the same overhang twice; (2) avoid palindromes; (3) no overhangs with the same three nucleotides in a row; (4) no more than two nucleotides in the same position; (5) avoid overhangs with either 0% or 100% GC.)
In fact, the NEB® team’s data showed that high-fidelity reactions could be done using overhang sets that broke rules 3—5. Fidelity, it turns out, might depend more on the individual sequence(s) than on how well that sequence fits into the five rules.
By 2020, they had used this data-optimized assembly design (DAD) to build out three tools to help researchers design high-fidelity Golden Gate Assemblies and overhang sets: NEBridge Ligase Fidelity Viewer™; NEBridge GetSet™; and NEBridge SplitSet™. With these tools, NEB successfully performed a 35-fragment Golden Gate reaction, using four-base overhangs, in a single tube. Their assembly had a predicted fidelity of 71% (Pryor et al., 2020). And in 2022, using the same tools, they successfully assembled a 40 kb T7 phage genome from 52 (!!) fragments in one very Golden (Gate) pot (Pryor et al., 2022).
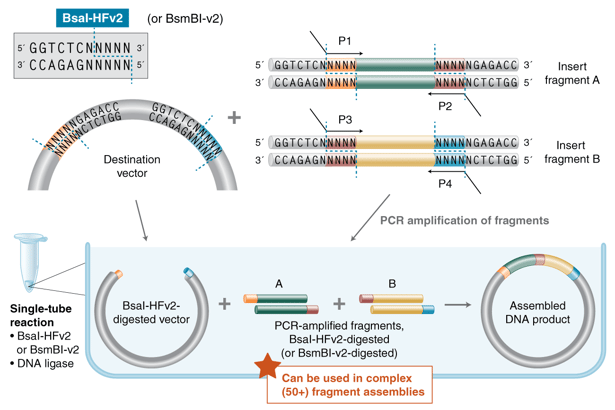
|
| Figure 1: Workflow for high complexity Golden Gate Assemblies. Image courtesy of New England Biolabs. |
DAD Tools
The three DAD tools used were key for ultra-high-complexity assembly. While the traditional rules of thumb for overhang set design allowed for high-fidelity assembly for sets of up to 10-12 fragments (Pryor et al., 2020), and randomly selected non-palindromic pairs allows for high-fidelity assembly for up to sets of up to 6-8 overhangs, the data-driven approach was needed to massively expand complexity. The three tools developed, outlined below, allow for researchers to select large sets (with overhangs that violate some of the traditional design rules) for successful high-complexity Golden Gate Assembly reactions.
NEBridge Ligase Fidelity Viewer
This tool allows a user to check fidelity of an overhang set by uploading the set with the Type IIS restriction enzyme and thermocycler protocol. It will also identify overhangs with a high potential for mismatches, so you can redesign individual pieces as needed to improve fidelity.
NEBridge GetSet
Need a set of overhangs, stat? GetSet can do that for you, complete with predicted fidelity. You can specify number of overhangs, three or four base-pair sequences, and exclude or include specific overhang sequences, allowing it to be used to expand already-existing sets. Note that this tool relies on a stochastic search algorithm rather than pre-designed sets, so identical searches can return multiple answers. You might want to take advantage of their save and recall functions when you get results you like.
NEBridge SplitSet
If you want to design an overhang set specific to your DNA sequence, SplitSet is the tool for you. Once you input a sequence, SplitSet finds optimal fusion sites to split it into multiple fragments. You can define the number of fragments, and the “search windows” you’d like the tool to use to identify fusion sequences for either a circular or linear assembly. If you aren’t constricted to a specific number of fragments, you can also ask it to give the maximum or minimum number of fragments, or defined split regions. SplitSet will then return the highest-fidelity overhang set it can find for those parameters. For even more specificity, you can exclude specific sequences or set a narrow search window for a site—or sites—that must be used.
Each of the tools works with either 3-base or 4-base overhangs, and with a variety of different enzymes and cycling conditions (see Table 1).
|
Enzyme |
Temperature |
Incubation time/cycling |
Overhang length |
|
T4 DNA ligase |
25 °C |
1 hour |
3-base or 4-base |
|
T4 DNA ligase |
25 °C |
18 hours |
3-base or 4-base |
|
T4 DNA ligase |
37 °C |
1 hour |
3-base or 4-base |
|
T4 DNA ligase |
37 °C |
18 hours |
3-base or 4-base |
|
Bbsl-HF®, 1x NEBridge® Ligase MM |
37-16 °C |
cycling |
4-base |
|
Bsal-HFv2® |
37 °C |
Static |
4-base |
|
Bsal-HFv2, 1x NEBridge Ligase MM |
37-16 °C |
cycling |
4-base |
|
Bsal-HFv2 |
37-16 °C |
cycling |
4-base |
|
BsmBI-v2 |
42-16 °C |
cycling |
4-base |
|
BsmBl-v2, 1x NEBridge Ligase MM |
42-16 °C |
cycling |
4-base |
|
BspQI, 1x NEBridge Ligase MM |
42-16 °C |
cycling |
3-base |
|
PaqCl®, 1x T4 DNA Ligase Buffer |
37-16 °C |
cycling |
4-base |
|
PaqCl, 1x NEBridge Ligase MM |
37-16 °C |
cycling |
4-base |
|
Sapl |
37-16 °C |
cycling |
3-base |
|
Sapl, 1x NEBridge Ligase MM |
37-16 °C |
cycling |
3-base |
Table 1: Enzyme and cycling conditions available in DAD tools
While DAD made an incredibly complex reaction possible, NEB also optimized the thermocycler protocols and made sure that T4 ligase was the correct choice for the job compared to T7 ligase (it was—see Potapov et al., 2018). More complex reactions benefited from longer incubation times and didn’t necessarily require cycling.
The Pros and Cons
Researchers used the tools above to complete a 52-piece assembly of a 40 kb T7 phage genome. They first compared 10-piece linear (the native state of the T7 genome) and circular assemblies. While Golden Gate Assembly worked for all the assembly types, circular assemblies yielded 500 times the number of plaques than the linear ones.
The 52-piece circular assembly of the T7 genome had lower efficiency than the 10-piece circular assembly, with the 52-piece approach yielding approximately ~800-fold fewer plaques than the 10-piece one. Though it was possible to select for correctly assembled phage genomes, it required a very stringent selection process and benefited from a longer reaction time of 15 hours. This suggests that 52 fragments may be near the upper limit of what can be accomplished with Golden Gate Assembly—even when getting help from DAD.
The 35-piece assembly mentioned above had a remarkably high 71% fidelity, while the 52-piece assembly of a lac operon dropped to 49% fidelity, with significant lot-to-lot variation. Additionally, this assembly, which used only antibiotic screening for selection, failed under normal cycling protocols, and required a 48-hour incubation at 37 °C. While the 52-piece reaction shows massively complex assemblies are possible, the two sets of data together indicate that 35-pieces is a reasonable upper limit for Golden Gate Assembly size.
Ultimately, data-driven Golden Gate Assembly greatly increases the potential complexity allowable by a favored, fairly easy cloning technique (which may be of particular interest to MoClo users!). If you’re interested in complex Golden Gate Assembly, a great place to start is the 24-piece lac operon assembly deposited by NEB – and don’t forget to ask DAD for some help!
References
Potapov, V., Ong, J. L., Kucera, R. B., Langhorst, B. W., Bilotti, K., Pryor, J. M., Cantor, E. J., Canton, B., Knight, T. F., Thomas C. Evans, J., & Lohman, G. J. S. (2018). Comprehensive Profiling of Four Base Overhang Ligation Fidelity by T4 DNA Ligase and Application to DNA Assembly. ACS Synthetic Biology. https://doi.org/10.1021/acssynbio.8b00333
Pryor, J. M., Potapov, V., Bilotti, K., Pokhrel, N., & Lohman, G. J. S. (2022). Rapid 40 kb Genome Construction from 52 Parts through Data-optimized Assembly Design. ACS Synthetic Biology, 11(6), 2036–2042. https://doi.org/10.1021/acssynbio.1c00525
Pryor, J. M., Potapov, V., Kucera, R. B., Bilotti, K., Cantor, E. J., & Lohman, G. J. S. (2020). Enabling one-pot Golden Gate assemblies of unprecedented complexity using data-optimized assembly design. PLOS ONE, 15(9), e0238592. https://doi.org/10.1371/journal.pone.0238592
Topics: Plasmids 101, Plasmid Cloning, Plasmids






Leave a Comment