CRISPR has greatly enhanced the ability of scientists to make genomic alterations, bringing about a revolution in genome engineering, with new techniques rapidly being developed. Performing a CRISPR experiment requires delivery of, at minimum, two components: the Cas9 protein and a guide RNA (gRNA) targeting your genomic site of interest. This is commonly performed by transfecting cells with a plasmid, such as PX459, which encodes Cas9 and contains a site for inserting a custom gRNA. While this methodology has proven to be incredibly valuable to scientists, there are some potential complications that must be considered when using this method:
- Cells must be amenable to transfection or viral transduction
- Appropriate promoters must be chosen for both Cas9 and gRNA expression
- Plasmid DNA may be incorporated into the genome
- Off-target effects can occur due to prolonged Cas9 expression
- The requirement for Cas9 transcription and translation delays editing
What are Cas9-gRNA ribonucleoproteins?
One alternative approach, which avoids many of these complications, is to directly deliver a ribonucleoprotein (RNP), consisting of the Cas9 protein in complex with a targeting gRNA, to your cells of interest. Cas9 RNPs are capable of cleaving genomic targets with similar efficiency as compared to plasmid-based expression of Cas9/gRNA and can be used for most of the current genome engineering applications of CRISPR including: generating single or multi-gene knockouts in a wide variety of cell types, gene editing using homology directed repair (HDR), and generating large genomic deletions.
What are the advantages of Cas9 RNPs?
There are many advantages to using RNPs for CRISPR experiments.
- The RNP method can often be used in cells that are difficult to transfect, such as primary cells.
- Using RNPs can also alleviate difficulties with protein expression that occur in cells where common eukaryotic promoters (such as CMV or EF1A promoters found in many CRISPR plasmids) are not expressed.
- Because this method does not require the delivery of foreign DNA, and the Cas9-gRNA RNP is degraded over time, using RNPs may limit the potential for off-target effects.
- Cas9 RNPs are detectable at high levels shortly after transfection, and are quickly cleared from the cell via protein degradation pathways.
- This makes Cas9 RNPs useful for CRISPR applications where limited expression of Cas9 is required and specificity is a concern, such as knockout generation or homologous recombination. However, experiments that require long-term expression of Cas9, such as visualizing genomic loci using fluorophore tagged dCas9 may require the use of plasmid or viral-mediated delivery.
You’ve decided to use RNPs, now what?
Preparing the Cas9-gRNA ribonucleoprotein
The first step in this type of CRISPR experiment is the generation of the RNP complex. While one option is to purchase the Cas9 protein and a gRNA from a commercial vendor, this can often be expensive. An alternative approach is to express and purify His-tagged Cas9 from E. coli using a plasmid such as pET-28b-Cas9-His from Alex Schier’s Lab. gRNAs can be in vitro transcribed from ssDNA, which can be generated by commercial vendors such as IDT. These two components are then incubated together to form the RNP. Detailed protocols outlining these steps have been made publicly available by both the Jacob Corn and Alex Schier laboratories.
Delivering Cas9-gRNA ribonucleoproteins
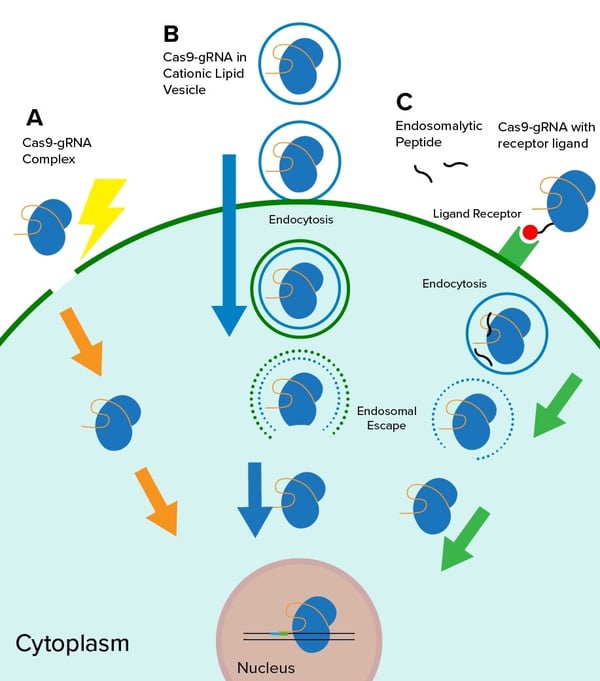
An advantage of performing CRISPR experiments using RNPs is the variety of methods that can be used to deliver a Cas9-gRNA RNP. This advantage has allowed scientists to perform in vitro and in vivo CRISPR studies in experimental systems that may not be amenable to plasmid based methods.
One of the most common techniques for delivery of RNPs is electroporation (A in the figure above), which generates pores in the cell membrane, allowing for entry of the RNP into the cytoplasm. In addition to the use of this technique in cell culture, it has also been applied to genome editing of mouse zygotes, through a process known as CRISPR-EZ (CRISPR RNP Electroporation of Zygotes) (Chen et al. 2016). For CRISPR experiments that involve HDR, electroporation can be combined with cell-type specific reagents in a technique known as nucleofection, which forms pores in the nuclear membrane, allowing for entry of a DNA template.
Other techniques, such as lipid-mediated transfection (B in the figure above), are in the early stages of development. David Liu’s Lab has demonstrated the use of cationic lipid-mediated methods to deliver Cas9-gRNA RNPs to hair cells in the mouse inner ear (Zuris et al. 2015). A more targeted approach using RNPs has recently been described in vitro, with the hope of eventually using this method for in vivo applications. This technique utilizes Cas9 proteins harboring receptor ligands (C in the figure above), which result in the internalization of Cas9-gRNA RNPs by specific cell types (Rouet et al. 2018). This was accomplished using a Cas9 mutant (Cas9M1C/C80S) that harbors two surface exposed cysteines, including the native C547, allowing for ligation to pyridyl disulfide-activated ligands.
Due to the concerns of off-target mutations and transgene integration in crops, delivery of Cas9-gRNA RNPs to plants, as an alternative to plasmid based techniques, has been an area of intense study. Two methods to accomplish this are Polyethylene Glycol (PEG) mediated transfection of protoplasts and biolistic bombardment of immature embryos (Liang et al. 2017). PEG-mediated methods, while technically less difficult to perform, often show low efficiency, while biolistic bombardment, which can have high efficiency, requires specialized equipment, such as a gene gun. Depending on the nature of the experiment, it is up to the researcher to determine which method is most appropriate for a given experiment.
Considerations for RNP-mediated CRISPR experiments
While using Cas9-gRNA RNPs for CRISPR experiments can reduce many potential complications, there are additional factors to take into account with these types of experiments. One important consideration is that the Cas9-gRNA RNP will be degraded by the cell over time. As a result, RNP mediated CRISPR experiments may not be suitable for experiments where stable Cas9 expression is required, such as when a Cas9 fusion to a fluorescent protein is used to label nucleic acid targets. While Cas9-gRNA RNPs show promise for in vivo studies, little is known about the immunogenicity and stability of the Cas9 protein in the host. Indeed, research has shown that a Cas9 protein from Geobacillus stearothermophilus is more stable in human plasma than the standard spCas9 (Harrington et al. 2017). Lastly, as with any scientific research, choice of a technique often comes down to the researcher’s familiarity with different systems. Using Cas9-gRNA RNPs in your experiments can require steps such as protein expression and purification, which will require additional laboratory reagents, equipment, and expertise.
Final thoughts
The Cas9-gRNA RNP method provides scientists with an additional means to deliver CRISPR components to their experimental systems of interest. Using this method can alleviate many issues that may arise when using plasmid based systems, most notably the ability to perform CRISPR studies in cells that are difficult to transfect or transduce. While using Cas9-gRNA RNPs may require the production of the Cas9 protein and a gRNA in your laboratory, once these have been acquired, genomic editing can be performed rapidly and, in many cases, with reduced chance for off-target effects. As the CRISPR field continues to evolve, it is likely that the use of RNPs will play an important role in its continued advancement.
 Andrew Hempstead is a Senior Scientist at Addgene. Andrew's interests include genome engineering and microbiology.
Andrew Hempstead is a Senior Scientist at Addgene. Andrew's interests include genome engineering and microbiology.
References
1. Chen, Sean, et al. "Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes." Journal of Biological Chemistry (2016): jbc-M116. PubMed PMID: 27151215. PubMed Central PMCID: PMC4938170.
2. Zuris, John A., et al. "Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo." Nature biotechnology 33.1 (2015): 73. PubMed PMID: 25357182. PubMed Central PMCID: PMC4289409.
3. Rouet, Romain, et al. "Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing." Journal of the American Chemical Society 140.21 (2018): 6596-6603. PubMed PMID: 29668265. PubMed Central PMCID: PMC6002863.
4. Liang, Zhen, et al. "Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes." Nature communications 8 (2017): 14261. PubMed PMID: 28098143. PubMed Central PMCID: PMC5253684.
5. Harrington, Lucas B., et al. "A thermostable Cas9 with increased lifetime in human plasma." Nature Communications8.1 (2017): 1424. PubMed PMID: 29127284. PubMed Central PMCID: PMC5681539.
Additional resources on the Addgene Blog
- Subscribe to CRISPR blog posts
- Learn about other CRISPR delivery techniques
- Find tips on gRNA design
Resources on Addgene.org
- Browse all CRISPR plasmids
- Find validated gRNAs
- Check out our CRISPR guide page
Topics: CRISPR, CRISPR 101, CRISPR Expression Systems and Delivery Methods






Leave a Comment