Read All Our Editor's Choice Blog Posts
The Addgene Blog has had quite the up-tick in views over the month of August - at over 56,000 views, we’ve grown 12% over last month! This was a bit surprising given how many scientists go on vacation in August (for another part of my job, I travel around the country and set up meetings with PIs to discuss the benefits of depositing… you’d be amazed by the number of out of office responses I’ve received). Clearly we have all of our writers to thank for the excellent content they’ve churned out in the last month.
 Google Forums Round Up: First Impressions of NgAgo
Google Forums Round Up: First Impressions of NgAgo
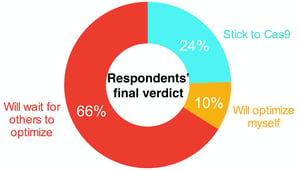 In particular, we need to thank Pooran Dewari who wrote this month’s most popular blog post on early user experiences with CRISPR alternative, NgAgo. As you might expect with any nascent technology, researchers are having trouble getting NgAgo genome editing up and running in their labs. Pooran surveyed a CRISPR google forum to get a better idea of how many people are having trouble, to determine if there have been any early successes, and to find out what delivery techniques researchers are using. The detailed results of his survey can be found in the full post but TLDR: many users have had difficulty with NgAgo, but a few successes have trickled into the survey. In response to a bit of a Twitter storm surrounding NgAgo, its creator, Han Chunyu of Hebei University of Science and Technology in Shijiazhuang, defended the technology in a news article in Nature. Our take home: users likely need more time to optimize and troubleshoot the system.
In particular, we need to thank Pooran Dewari who wrote this month’s most popular blog post on early user experiences with CRISPR alternative, NgAgo. As you might expect with any nascent technology, researchers are having trouble getting NgAgo genome editing up and running in their labs. Pooran surveyed a CRISPR google forum to get a better idea of how many people are having trouble, to determine if there have been any early successes, and to find out what delivery techniques researchers are using. The detailed results of his survey can be found in the full post but TLDR: many users have had difficulty with NgAgo, but a few successes have trickled into the survey. In response to a bit of a Twitter storm surrounding NgAgo, its creator, Han Chunyu of Hebei University of Science and Technology in Shijiazhuang, defended the technology in a news article in Nature. Our take home: users likely need more time to optimize and troubleshoot the system.
 Site Directed Mutagenesis by PCR & Single Base Editing with CRISPR
Site Directed Mutagenesis by PCR & Single Base Editing with CRISPR
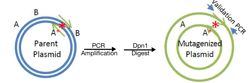 For my editor’s choice picks this month, I’m picking two posts that describe methods to make single base edits in DNA. The first, contributed by guest blogger Kristian Laursen from Cornell University, is a bit old school and provides a thorough walkthrough of how you can use PCR to make small sequence changes to plasmid DNA. This an oldie but a goodie, and in my personal experience, is a quick way to generate many mutated plasmid constructs.
For my editor’s choice picks this month, I’m picking two posts that describe methods to make single base edits in DNA. The first, contributed by guest blogger Kristian Laursen from Cornell University, is a bit old school and provides a thorough walkthrough of how you can use PCR to make small sequence changes to plasmid DNA. This an oldie but a goodie, and in my personal experience, is a quick way to generate many mutated plasmid constructs.
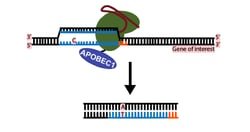 The second post was contributed by Addgene Scientist Mary Gearing and discusses a newer, CRISPR-based technique to generate C->T or G->A transitions in a target sequence in live cells. This technique uses catalytically inactive CRISPR endonuclease (dCas9) fused to a deaminase to make the transitions at target sequences specified by gRNAs. Caveats, potential specificity issues, and other means of generating mutations in live cells are all discussed further in this informative post. Interestingly, this technology may make some uses of Site Directed Mutagenesis by PCR obsolete!
The second post was contributed by Addgene Scientist Mary Gearing and discusses a newer, CRISPR-based technique to generate C->T or G->A transitions in a target sequence in live cells. This technique uses catalytically inactive CRISPR endonuclease (dCas9) fused to a deaminase to make the transitions at target sequences specified by gRNAs. Caveats, potential specificity issues, and other means of generating mutations in live cells are all discussed further in this informative post. Interestingly, this technology may make some uses of Site Directed Mutagenesis by PCR obsolete!
Honorable mentions
Of course we’ve had many other fantastic posts throughout the month including a first of its kind for the Addgene blog - Addgene Software Engineer Kris Shamloo wrote a very interesting post about how we develop our in house lab management software. The post is written in terms any programming noob (like myself) can understand. You’ll also find actionable advice on how to give a great science talk from Addgene Executive Director Joanne Kamens. I highly recommend taking a look at all of our posts as each one has something great to offer, I just need to get back to emailing (hopefully non-vacationing) scientists. You can find all of our August posts in chronological order here:
- Site Directed Mutagenesis by PCR
- Google Forums Round Up: First Impressions of NgAgo
- CRISPR Between the Genes: How to Experiment with Enhancers and Epigenomics
- 5 Tips for Troubleshooting Viral Transductions
- Single Base Editing with CRISPR
- Cas9 Activators: A Practical Guide
- 10 Steps to a Perfect Science Talk
- Developing Lab Management Software for Biology
- Adapting Toehold Switches to Detect Zika Virus
As always, if you have any questions or suggestions for us, feel free to comment below.
Happy reading!
Tyler J. Ford
Topics: Addgene News







Leave a Comment