 With the meteoric rise of CRISPR technology, the ability to direct enzymes – from nucleases to transcription factors – to specific sequences of DNA has become commonplace. This ability has opened up a world of possibilities in the engineering of complex gene networks. A comparable system for targeting specific sequences of RNA is highly desirable for extending the complexity of genetic circuits, allowing for tighter spatio-temporal control of gene expression within a cell. Thanks to the work of Huimin Zhao and colleagues, we now have just the tool…designer PUF proteins!
With the meteoric rise of CRISPR technology, the ability to direct enzymes – from nucleases to transcription factors – to specific sequences of DNA has become commonplace. This ability has opened up a world of possibilities in the engineering of complex gene networks. A comparable system for targeting specific sequences of RNA is highly desirable for extending the complexity of genetic circuits, allowing for tighter spatio-temporal control of gene expression within a cell. Thanks to the work of Huimin Zhao and colleagues, we now have just the tool…designer PUF proteins!
A newly available PUF Assembly Kit makes it possible to devise RNA binding proteins to hit any target of interest. The new tool was developed and implemented by applying the Golden Gate cloning method to human proteins known as Pumilio/fem-3 mRNA binding factors (PUF). In a single step, researchers can now assemble designer PUF domains for RNA specificity engineering.
“The RNA binding domain is interesting because by changing certain amino acids you can change the specificity,” explained Zhanar Abil of the University of Illinois at Urbana-Champaign.
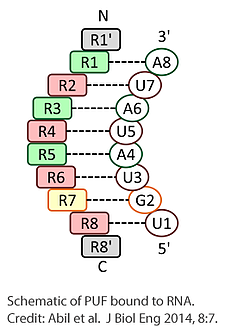
The PUF proteins include eight modular repeats, each of which binds a consecutive base in its RNA target. By changing amino acids, it’s possible to produce a protein that binds an alternate sequence. By comparison, Abil explained, the specificity of other RNA binding domains depends on the secondary structure of RNA. As a result, targeting the RNA of interest requires that the RNA be tagged.
“This tool makes it possible to make the protein specific to any endogenous RNA without tagging,” she said. “It opens a new avenue of modifying and manipulating endogenous eukaryotic RNA.”
Endless possibilities with PUF proteins
The tool she and her colleagues led by Zhao described and demonstrated in a report published in The Journal of Biological Engineering in March is fused to a translational repressor. In principle, however, designer PUF proteins can be fused to other components to lend them many other functions for use in RNA splicing, imaging, and more. Abil says she is currently working on a version that she hopes will change the intracellular localization of RNA.
“You can think of changing bases to other RNA bases or unnatural bases,” she said. “The possibilities, I think, are endless.” Abil says the new tool might even find therapeutic application in addition to their use in basic biology, for example, by targeting pathways underlying cancer or other disease processes.
Tips and tricks
For those interested to try the PUF Assembly Kit out for themselves, she does have a couple of tips: First, she recommends using standard BsaI in place of the high fidelity version (despite what the original paper says). In their hands, BsaI-HF has produced less reliable results.
She also notes that for reasons that are not yet entirely clear some PUF proteins lose their solubility. Her lab is currently investigating the issue and hopes to have a better understanding of the phenomenon soon.
Rest assured, as the PUF protein design tools continue to evolve and improve, Abil says they will make those new plasmids available through Addgene, and they hope others will do the same.
“This isn’t the final version,” she said. “If we learn how to make the library even better, we would be very happy to share updated versions of the kit.”
Reference
Abil Z, Denard CA, Zhao H. Modular assembly of designer PUF proteins for specific post-transcriptional regulation of endogenous RNA. J Biol Eng. 2014 Mar 1;8(1):7. doi: 10.1186/1754-1611-8-7. PubMed.
Related Resources
Topics: Other Plasmid Tools, Plasmids






Leave a Comment