This post was contributed by Greg Dingle, a software engineer with the Chan Zuckerberg Initiative.
We hereby announce the general availability of new a tool for CRISPR scientists––CrispyCrunch! CrispyCrunch is a web app that helps scientists design and analyze batches of CRISPR samples.
We invite you to jump in and try it out, or take a look at our live examples: experiment or analysis. In the rest of this article, we'll explain the thinking behind the tool, its key features, how it works, and how to use it.
Background on CrispyCrunch
 CrispyCrunch was initially built for the CZ Biohub project by Manuel Leonetti to tag all 22,000 protein coding genes in the human genome with fluorescent proteins. For each gene, we needed to design guide RNAs, donor templates and sequencing primers for quality control. At such a scale, automation is crucial, both for speeding things up and for standardizing the process.
CrispyCrunch was initially built for the CZ Biohub project by Manuel Leonetti to tag all 22,000 protein coding genes in the human genome with fluorescent proteins. For each gene, we needed to design guide RNAs, donor templates and sequencing primers for quality control. At such a scale, automation is crucial, both for speeding things up and for standardizing the process.
When we started, too much time for this project was spent selecting guides, designing primers, and running analyses. New team members have taken up to a month to design a single 96-well plate for HDR that satisfy all requirements.
To alleviate this bottleneck, we began building CrispyCrunch in July 2018 and in November, we began using it internally.
Guiding principles behind CrispyCrunch
While CrispyCrunch may evolve over time, you can depend on it to stick to a few guiding principles that we've followed since the beginning.
- It's free and open to all. The project is funded by CZI and the CZ Biohub for the acceleration of science.
- It's open-source. You can see all the current code on GitHub, ask us questions about it, or send us a pull request for improving it.
- As much as possible, it uses standard tools and databases, including primer3, biopython, bwa, samtools, bowtie, UCSC genome browser, Ensembl, and others.
- It builds on existing popular web services, currently Crispor and Crispresso, that adhere to the same principles listed here.
- It employs biological best practices. For example, it ranks guides by CFD score, and it mutates HDR donor templates to avoid re-cutting when needed.
- It gives you control over your data. CrispyCrunch keeps a record of all your designs and analyses, but it also allows you to download any generated info for your own purposes, or delete the originals if desired.
- It works as a whole or in part. For example, you can analyze experiments designed outside CrispyCrunch, or you can input pre-existing guides for primer design.
The ecosystem of CRISPR tools
Before deciding to build our own tool, we looked hard at existing tools. As has been written here on Addgene, there has been a Cambrian-like explosion of software tools accompanying the CRISPR revolution. (See omictools for even more.)
However, none of the tools we found performed batch guide design, batch primer design or batch analysis. We wanted to relieve scientists of the burden of having to manage all the standard information that goes into a 96-well plate and comes out of it. Biotech companies are known to have tools that do this internally, but they are not available publicly.
Furthermore, we did not find any tools which were optimized for HDR, except for TagIn. HDR requires the additional step of mutating guide sequences in a way that prevents re-cutting but does not disrupt gene expression. (See section below.)
For guide design, we decided to adapt the popular guide design tool Crispor by Max Haeussler. With a few extra lines of code, we enabled it to return guides and primers in batch, optimized for HDR.
Compared to wealth of resources for guide design, there are few tools to analyze CRISPR repair outcomes, which is important for quality control. The only ones we found that worked with NGS data were:
We chose to adapt Crispresso by Luca Pinello and Kendell Clement for batch analysis because of its ease-of-use and comprehensive reports. With CrispyCrunch, you get the all information from Crispresso plus a summary per batch.
Ranking guides for HDR in-depth
In other tools, guides are ranked by specificity of targeting or by cut-to-insert distance for HDR. (The notable exception is TagIn––see their "summarisation score".) For CrispyCrunch, we wanted to consider both factors, as a bench scientist would looking at a genome by eye. We came up with a ranking formula that does just this:
Rank = (compressed CFD score) * (gaussian normalized cut-to-insert distance)
This gives each guide a score ranging between 0 and 1.
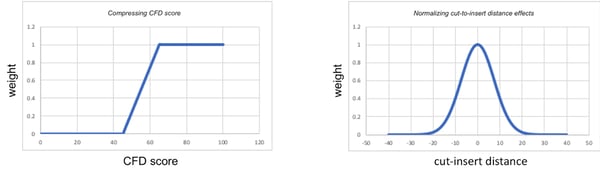
The cutting frequency determination (CFD score) is used to measure specificity of a guide to the target. The CFD score ranges between 0 and 100 for each guide, with 100 being the strongest interaction between the guide and the target and 0 being the weakest interaction due to mismatches between the guide and the DNA target. CFD scores are then modified (“transformed”) such that weak interactions (e.g., scores <20) are reduced to 0 and strong interactions (e.g., scores >80) are transformed to 100. This transformation (illustrated in the graph below) is performed because weak gRNAs (CFD < 20) bind almost not at all, and strong gRNAs (CFD > 80) bind almost as much as perfect gRNAs (CFD = 100). CFD scores (which range between 0 and 100) are then converted to a weight (which ranges between 0 and 1) so that they can be used in the rank equation above.
Cut-to-insert distance was fitted to experimental data of its effect on HDR efficiency. These values (ranging from -40 bp to 40 bp) are also converted to a weight (which ranges between 0 and 1) (shown in the graph below) so that it can be used in the rank equation above. Generally, the greater this distance between the cut and the insert, the more poorly the guide performs HDR.
Optimal mutation in-depth
One the most sophisticated features of CrispyCrunch is how it mutates guide sequences to prevent re-cutting of the sequence inserted by HDR. Although this practice is generally advised, there is little detail published on how to do it best.
Scientists are typically advised to mutate the PAM to which the guide binds. However, if the PAM resides in a coding region, the mutation may cause side-effects. Further, work by John Doench et al. shows that mutating a single base pair may not be enough to prevent re-binding. Lastly, the sequence inserted by HDR may inadvertently re-create a match for the active guide.
With these constraints in mind, we implemented the following algorithm for optimal mutation in CrispyCrunch:
- If the guide PAM resides outside a coding region in a UTR, CrispyCrunch flips the two important base pairs of the PAM (NGG -> NCC, CCN -> GGN). This ends the mutation process.
- If the guide PAM is within the coding region, the algorithm then compares the guide sequence to every possible 23bp sequence (protospacer + PAM) in the target region after HDR. If the max CFD score is less than 0.03, it stops mutating. (This happens often because the HDR inserted sequence splits up the guide sequence.)
- If neither of the above are options, the algorithm silently mutates codons in the guide sequence, one by one, from the PAM outwards so that the DNA sequence is altered but the protein sequence they encode is not. After each mutation, it will check the CFD score. If it is below 0.03, it stops mutating. (Note: the most common synonym in the human genome is chosen for a silent mutation.)
How to use CrispyCrunch
CrispyCrunch take you through the whole process from experimental design to analyzing results. To use this tool, simply follow these steps:
- Create a new experiment in the program
- Design gRNAs by inputting the target regions
- Design donor RNA
- Design primers
- Review and order reagents
- Perform your wet lab experiment
- Analyze the data
Conclusion
CrispyCrunch is now publicly-available for high-throughput design and analysis of CRISPR experiments. It has certainly boosted our productivity in the Biohub. We hope it will do the same for you.
Send us your questions or comments to gdingle@chanzuckerberg.com. We're eager to hear your thoughts.
Many thanks to our guest blogger, Greg Dingle!
 Greg Dingle is a software engineer with the Chan Zuckerberg Initiative, working on assignment at the Biohub in San Francisco. Before starting his career in software, he completed a MSc in Psychology at McMaster University in Canada. He's happy to be contributing again to science.
Greg Dingle is a software engineer with the Chan Zuckerberg Initiative, working on assignment at the Biohub in San Francisco. Before starting his career in software, he completed a MSc in Psychology at McMaster University in Canada. He's happy to be contributing again to science.
Acknowledgements
- Chan Zuckerberg Initiative
- Chan Zuckerberg Biohub
- Manuel Leonetti
- Ryan Leenay
- Jason Li
- Max Haeussler
- Luca Pinello
- Kendell Clement
- Andy May
Additional resources on the Addgene blog
- Read our CRISPR 101 blog posts
- Learn about some considerations when designing gRNAs
- Find other CRISPR software
Resources on Addgene.org
- Find CRISPR plasmids and resources
- Download the CRISPR eBook
- Read our CRISPR guide
Topics: CRISPR, Other CRISPR Tools





Leave a Comment