Last updated Apr 17, 2020.
 Before being adapted by scientists to edit the genome of virtually any organisms on this planet, CRISPR-Cas systems were merely adaptive immune systems that provide bacteria protection against infectious agents. Several enzymes behind this immunity have already been discovered and studied but it is only the tip of the iceberg as it has been predicted that many others are still unknown.
Before being adapted by scientists to edit the genome of virtually any organisms on this planet, CRISPR-Cas systems were merely adaptive immune systems that provide bacteria protection against infectious agents. Several enzymes behind this immunity have already been discovered and studied but it is only the tip of the iceberg as it has been predicted that many others are still unknown.
Identifying Cas14 variants
In this quest to discover new and maybe more efficient Cas systems, Jennifer Doudna’s lab analyzed metagenomic datasets to try to determine whether simpler and maybe smaller Cas systems could exist in nature.
To do this, they mined a database of microbial genome and metagenome from the Department of Energy’s Joint Genome Institute for uncharacterized genes that were proximal to a CRISPR array and the universal CRISPR integrase, cas1. They discovered a family of CRISPR-Cas systems containing cas1, cas2, cas4, and a new gene cas14. cas14 encodes for a small Cas protein (40-70 kDa), which is half the size of other Cas proteins found in the so-called class 2 CRISPR-Cas systems.
There are 24 variants of the cas14 gene that cluster into 3 subgroups (cas14a-c). All these variants share the presence of a predicted RuvC nuclease domain, characteristic of CRISPR-Cas enzymes. In contrast to other Cas enzymes, Cas14 has not been found in bacterial genomes but only in the genome of a group of Archaea. Consistent with this origin, the authors suggest that Cas14 could be a more primitive version of the larger and more complex Cas9 and Cas12 proteins.
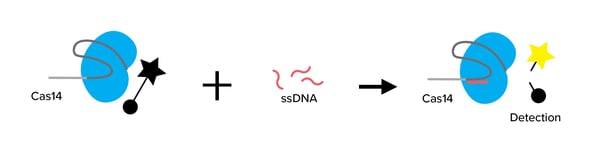
Cas14 cleaves ssDNA
But what is the function of this new system? And could it be a new tool for genome editing? To test the potential of Cas14, the Doudna lab cloned the cas14 gene together with the adjacent CRISPR array and intergenic regions containing the putative tracrRNA into a plasmid and expressed it in an E. coli bacteria strain. They found that Cas14 could bind and cleave a targeted sequence of ssDNA. Unlike Cas9, Cas14 does not require a PAM sequence. In addition to this cis cleavage, the authors showed that Cas14 could indiscriminately cleave ssDNA in trans like Cas13 and Cas12 do for RNA and dsDNA, respectively. However the authors found that Cas14 is much more specific in its recognition of ssDNA than Cas13 or Cas12 are for the other types of nucleic acids and required a certain sequence specificity in the middle of the gRNA to be activated.
Cas14 in diagnostics
By cutting ssDNA but not dsDNA Cas14 is not a good candidate for a new genome-editing tool but it could be a fantastic add-on to a diagnostic toolkit, called DETECTR, which has been created by the authors. This kit uses Cas12 and Cas13 to quickly detect the presence of infectious organisms (ex: SARS-CoV-2) and genetic mutations. By providing a gRNA specific of a viral sequences, for instance, the enzymes are activated and can cleave fluorescent probes of dsDNA and RNA in trans. Fluorescent signal indicates the presence of this viral sequence in the assay.
By adding Cas14 to the mix, you get the ultimate combination enabling the detection of RNA, dsDNA, and now ssDNA. By comparing the ability of Cas12 and Cas14 to detect SNPs using the DETECTR kit, the authors showed that improved specificity of Cas14 enables high-fidelity SNP genotyping. The development of Cas14-DETECTR will soon enable the quick and efficient diagnoses of infections, cancer, and other diseases.
References
1. Harrington, Lucas B., et al. "Programmed DNA destruction by miniature CRISPR-Cas14 enzymes." Science 362.6416 (2018): 839-842. PubMed ID PMID: 30337455.
Additional resources on the Addgene Blog
- Learn more about DETECTR
- Browse all of the CRISPR blog posts
- Download our CRISPR cheat sheet
Resources on Addgene.org
- Find the Cas14 plasmids here
- Checkout our CRISPR guide page
- Browse the CRISPR collection

Topics: CRISPR, Cas Proteins





Leave a Comment