Have you ever been stuck googling about an antibody that will be essential to your new research project, but unsure which one will mark your target the best, or even which one will work for your application? Like many scientists, I have spent hours sifting through comments and papers to find the most reliable (and cost-effective) antibody for my experiment – the “goldilocks antibody.”
Antibodies are one of the most commonly used research tools in scientists’ pockets. They allow researchers to identify and isolate very specific biological targets. However, as popular and useful antibodies are, they are in a midst of a crisis. The “antibody crisis” came to light as antibodies became commercialized, revealing the unreliability of antibodies. There are three main underlying causes of this unreliability: cross-reactivity (binds to non-target proteins), variability (separate batches can perform differently – happens when the antibody is produced from a new set of animals), and wrong application (different experiments/conditions can change binding ability – i.e., the antibody can no longer bind to the target). Unreliable antibodies cause research that cannot be reproduced and projects that cannot be completed. A 2015 review analyzing and explaining the antibody crisis states that up to 50% of all commercially available antibodies are unreliable (Baker, 2015), causing a huge waste of precious samples, time, and funds – up to US$350 million in 2015 alone and probably more today! – (Bradbury and Plückthun, 2015). To avoid being caught in the antibody crisis, one must corral and corner the goldilocks antibodies.
A goldilocks antibody is highly specific (minimal binding to off-target molecules), reliable (wouldn’t perform differently over time), compatible with the desired applications (such as immunohistochemistry (IHC) or western blot), and cost-effective. If only there was an open-source website that contained all the goldilocks antibodies in one place... well, lucky for you one is in the making by Dr. Zhuhao Wu and his colleagues!
Dr. Wu and his lab members (like me!) are dedicated to creating a public and accessible database for low-cost, high-quality antibodies for IHC. Working with collaborators and support from the Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative Cell Census Network, the Wu lab will be screening over 3,000 antibodies for use in mice and human brain IHC and uploading verified antibodies onto the mAb3D atlas website. This website can then be used by thousands of scientists to confidently choose reliable and consistent antibodies for specific markers.
The Screening Criteria
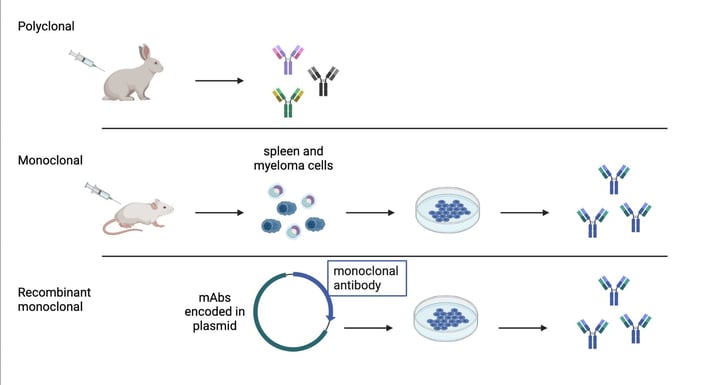
While choosing which antibodies to screen, we kept in mind our own lab’s purposes. The Wu lab mostly uses whole-mount IHC techniques on rodent brains and is expanding to more complex brain systems such as non-human primates and humans. Since we need larger-volume antibodies for thicker tissues and whole brains, we chose monoclonal antibodies (mAbs) and their recombinant forms (rAbs). mAbs and rAbs have higher homogeneity than polyclonal antibodies because of their immunoglobin component that can be produced in large “copies.” This homogeneity circumvents the batch-to-batch variability found in polyclonal antibodies, creating a potentially unlimited supply of identical antibodies (See Fig 1.). rAbs are a relatively new technology that takes advantage of DNA sequencing to turn mAb clones into expression constructs to be conveniently modified and engineered.
|
|
| Fig. 1: Antibody production methods |
NeuroMab and others have an extensive library of monoclonal antibodies; Addgene collaborates with these companies to create and provide general availability of their recombinant antibodies. Since recombinant antibodies are a newer technology, it is important to show through qualitative data that they are a long-term solution to reproducibility and cost-effectiveness for broad and large quantity uses. We therefore use both rAb and mAB in our screening.
To screen the antibodies, we have developed a criterion that includes accuracy, strength, and reproducibility. We test the antibodies on human and mouse tissue and compare the IHC results to both NeuroMab and the Allen Brain Atlas. The comparison with two atlases enables us to see the reproducibility and accuracy while screening. We also compare different clones of the same target to see if the results are the same/similar, emphasizing the antibody’s specificity.
Strength and accuracy
The strength is based on binding specificity and exposure time. We first have a negative control which does not have primary and only contains secondary antibodies, allowing us to only image autofluorescence i.e., background noise. We then compare the negative control to a sample with the primary included to confirm that the signal is from the primary antibody and not from the background noise. If the sample images similarly to the negative control, that informs us that the primary antibody is very weak or with general non-specific binding. If the sample differs from the negative control, such as showing a very bright signal only in the hippocampus compared to other regions, that informs us that this antibody has specific binding and has a strong signal. We call the unique pattern of the antibody binding “distinct imaged pattern.” The distinct imaged pattern should align with what the atlases (mentioned above) and literature suggest, providing an expectation and comparison on the antibody signal’s location. If a distinct imaged pattern does NOT align with the atlases/literature and is unrelated to the target protein, we categorize the antibody as having non-specific binding in our IHC condition.
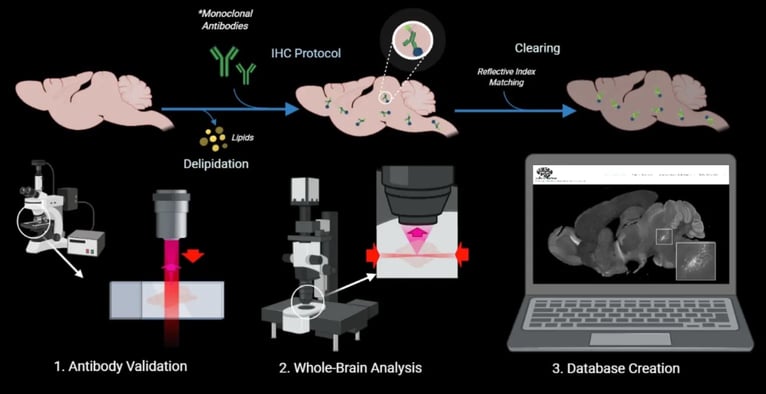 |
| Fig. 2: Schematic of the goldilocks antibody selection process |
The exposure time of the primary antibody compared to negative control also determines the signal’s strength. Exposure time is the amount of time the sample needs to be under light to discern the signal. If the exposure time is long (e.g. > 1/7s) then the signal is weak and vice versa for a strong signal. However, exposure time and its relationship to signal depend on what you’re comparing your sample to. For example, we use our negative control to compare exposure times of our samples. A prominent, strong signal has a shorter exposure time than the negative control, and a weak signal has a longer exposure time closer to the negative control.
Reproducibility
In order to attain non-bias assessment, multiple people in the lab conduct “independent screening”, where they look at the same section and determine the antibody’s reliability through their own analysis of the background noise, distinct imaged pattern, and exposure times. To test the antibody’s reproducibility, each antibody is independently screened twice, confirming it will produce the same results even to a different sample. Finally, as a lab we discuss the results together, explaining our reasoning to determine if the antibody is negative (poor quality) or positive (good quality). If the antibody meets these criteria and deemed positive, it will be released through the website for public reference.
The Gift of Giving
As an open-source database, the mAb3D Atlas will foster collaboration, progress, and innovation by allowing scientists to find and utilize reliable, high-quality antibodies. We are creating a database for goldilocks antibodies and, alongside that, a publicly available 2D/3D mouse atlas from antibodies that we screen. This atlas will enable scientists to have an intuitive guide for checking their staining and neuroanatomy without the fear of referencing a wrong section. The antibody screening and development of a focused reference bring us one step closer to resolving the antibody crisis. The mAb3D Atlas is a public service to the science community, and I am excited to be a part of it! If you are interested to get involved by submitting data, sending antibodies, and/or working with Dr. Wu please contact us here!
Ava Shipman is a research assistant in lab of Zhuhao Wu at the Icahn School of Medicine at Mount Sinai.
References and resources
References
Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015 May
21;521(7552):274-6. doi: 10.1038/521274a. PMID: 25993940.
Bradbury, A., Plückthun, A. Reproducibility: Standardize antibodies used in research. Nature 518, 27–29 (2015). https://doi.org/10.1038/518027a
Freedman LP, Cockburn IM, Simcoe TS (2015) The Economics of Reproducibility in Preclinical
Research. PLoS Biol 13(6): e1002165. https://doi.org/10.1371/journal.pbio.1002165
More resources on the Addgene blog
Antibodies 101: Validation
Selecting the Right Antibody
Plasmid-based Recombinant Antibodies
Topics: Antibodies






Leave a Comment