Addgene has recently announced the launch of an open-access recombinant antibody resource called NABOR (Neuroscience AntiBody Open Resource). The antibodies distributed through NABOR are recombinant antibodies expressed from plasmids that have been deposited with Addgene. Here, we take a closer look at the construction and validation of some of these tools created by the Trimmer Lab that will be the first antibodies distributed by Addgene. The plasmids used to generate these antibodies were based off of a large collection of mouse monoclonal antibodies (mAbs) that had already been thoroughly validated. The creativity in employing a variety of cloning techniques and troubleshooting methodologies drew me to learn more about the process that was used to create this important collection.
Creating the Plasmids
To begin constructing the plasmids for expression of mAbs, the Trimmer lab isolated RNA from cryopreserved hybridomas, immortalized cells created by fusing an activated B cell that produces the antibody of interest with a myeloma cell. This RNA was then used to generate cDNA, which was used as a template for PCR amplification with a degenerate primer set to amplify IgG variable light (VL) and variable heavy (VH) chain sequences. Fusion PCR was performed to create an amplicon that was cloned into a plasmid backbone for expression of the mAb heavy and light chain, each from a constitutively active CMV promoter. Constructs were then verified by both diagnostic restriction digest and colony PCR.
.jpg?width=750&name=elife-43322-fig1-min%20(1).jpg)
|
| Fig. 1: (A) Schematic of cloning, expression and validation pipeline. (B) Schematic shows the separate elements of the R-mAb expression plasmid involved in coexpression of light (green) and heavy (blue) chains as driven by two CMV promoters (orange). Figure from Andrews, et. al., Elife |
A limitation of hybridoma generated mAbs is that if the frozen cell line becomes non-recoverable, the stock of the mAb becomes limited, which can be a threat to future reproducibility. The Trimmer Lab found that in addition to recovering RNA from hybridomas grown in tissue culture, they were also able to isolate RNA from non-recoverable hybridoma stocks, without having to grow up the cells. This RNA could then be used to generate functional recombinant monoclonal antibody (R-mAb) expression plasmids, allowing for important reagents that could have been lost to continue to be available to the research community.
Validation
For most cloning projects, the next step would be to validate your new construct by Sanger sequence analysis. But for this research, it was likely that the sequence of the functional VH and VL sequence was not known for each of the mAbs, given the investment it would take to WGS the lab’s entire hybridoma collection. Instead, the Trimmer Lab moved to direct functional validation by immunofluorescence-immunocytochemistry (IF-ICC.) This method was chosen because cells transfected with a plasmid to express the target antigen could be screened in a high-throughput manner. Cells were labeled with both a hybridoma generated mAb and the R-mAb, with the R-mAb being verified when the labeling matched that which was observed for the hybridoma mAb.
While screening by IF-ICC worked well for identification of the correct clones, it was determined that for some constructs more than 90% of the colony PCR validated clones unexpectedly failed to produce functional R-mAbs. It was recognized that an aberrant IgG light chain transcript was expressed by some of the hybridomas and, as a result, subsequently cloned into the expression constructs. The Trimmer lab found that the recognition sequence for the restriction enzyme BciVI was present in the aberrant light chain sequence, but was predicted to occur at a low frequency within functional light chain sequences. By performing a BciVI digest of the PCR products prior to cloning, which in most cases left the functional light chain intact, the authors were able to greatly increase the number of functional constructs that were created. This greatly limited the number of clones that needed to be screened through functional assays.
Following functional verification, the R-mAb expression plasmids were DNA sequenced to determine the VH and VL sequence. These plasmids have been deposited with Addgene for distribution to the scientific community with the plasmid sequences available from each plasmid webpage. Importantly, the methods developed by the Trimmer Lab for construction and validation of R-mAb expressing plasmids could be used by other labs to preserve and share their own collections.
Why Recombinant Antibodies?
One of the benefits of using a plasmid based system for expression of R-mAbs is that it can easily be engineered. Like humans, mice express multiple subclasses of IgG antibody. Broad range secondary antibodies are commonly used in research laboratories to detect the different subclasses, but there are also secondary antibodies that are specific to each subclass. Approximately 70% of mouse mAbs are IgG1 subclass (Manning et al. 2012), which limits the use of subclass specific secondary antibodies to detect multiple targets with different mAbs in the same experiment. The Trimmer Lab found that they could modify the CH encoding sequence within the plasmid backbone to generate R-mAbs of different subclasses, including subclasses that were not produced by the original hybridoma. By using subclass specific secondary antibodies and subclassed switched R-mAbs, multiple targets could now be visualized in a single experiment, which had not been previously possible. This advancement could be especially exciting for scientists working to visualize multiprotein complexes.
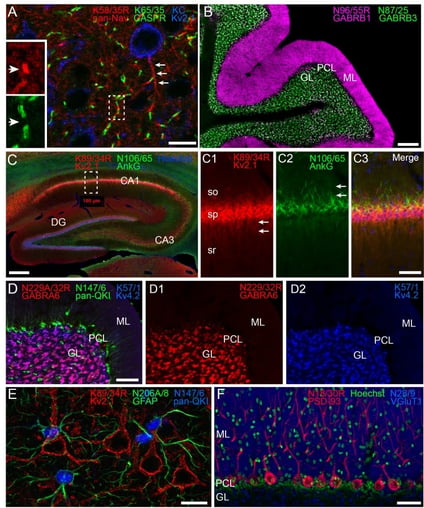 |
| Fig. 2. Multiplex immunolabeling with subclass-switched recombinant antibodies in adult rat brain. Figure from Andrews, et. al, Elife. Detailed figure legend here. |
The Trimmer Lab has continued to work to make the sequences of monoclonal antibodies publicly available. The Neuro Mab Sequencing Initiative uses high-throughput DNA sequencing to determine the VH and VL sequences from the Trimmer lab hybridoma collection and make them publicly available. The lab has also expanded their recombinant affinity reagent tools to include nanobodies. At Addgene, we are excited to be part of this initiative and to continue to accelerate research and discovery by improving access to useful research materials and information.
References and Resources
Additional Resources on the Addgene blog:
Plasmid-Based Recombinant Antibodies
Plasmids 101: Secondary Nanobody Toolbox
Antibodies 101: Introduction to Antibodies
Resources on Addgene.org
Browse Addgene's recombinant antibody plasmid collection
References
Andrews NP, Boeckman JX, Manning CF, Nguyen JT, Bechtold H, Dumitras C, Gong B, Nguyen K, van der List D, Murray KD, Engebrecht J, Trimmer JS. A toolbox of IgG subclass-switched recombinant monoclonal antibodies for enhanced multiplex immunolabeling of brain. Elife. 2019 Jan 22;8:e43322. doi: 10.7554/eLife.43322. PMID: 30667360; PMCID: PMC6377228.
Manning CF, Bundros AM, Trimmer JS. Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS One. 2012;7(6):e38313. doi: 10.1371/journal.pone.0038313. Epub 2012 Jun 1. PMID: 22675541; PMCID: PMC3365890.
Topics: Antibodies






Leave a Comment