Nanobodies from camelids have gotten a lot of press for being a promising approach for treatment or prophylaxis of COVID-19. Researchers identified nanobodies that bind to coronavirus spike proteins (Wrapp et al., 2020), which sit on the surface of coronaviruses and enable the virus to enter host cells. To do this, they injected the spike protein from two different coronaviruses into a llama over several days and isolated special camelid antibodies that bind to the spike proteins in cell culture, and inhibit these viruses.
But llama immunization has its drawbacks: it requires a camelid animal facility which can be expensive to gain access to routinely, and it can be especially challenging when trying to develop antibodies against unstable targets like membrane proteins that may unfold due to camelid high body temperature (37.2 - 39.2°C). The Seeger lab found a way around this by using plasmids to generate sybodies, synthetic nanobodies against the spike protein (Walter et al., 2020). No llamas required.
The advantages of synthetic nanobodies
Nanobodies are small (12-15 kDa) single-domain proteins that can bind effectively to protein targets, similar to how antibodies do. The structure of the common antibody is heftier (~150 kDa) and Y-shaped because it contains two large heavy chains and two small light chains that stick out to form the “arms” holding the highly variable regions that bind to specific antigens. In contrast, nanobodies are a single heavy domain fragment derived from a camelid antibody called a heavy chain antibody (HCab). Because of their small size, nanobodies can be synthetically expressed off of plasmids in bacteria, and they can target epitopes that are hard for antibodies to get to, like ones that are hidden in the molecular crevices proteins on the cell surface.
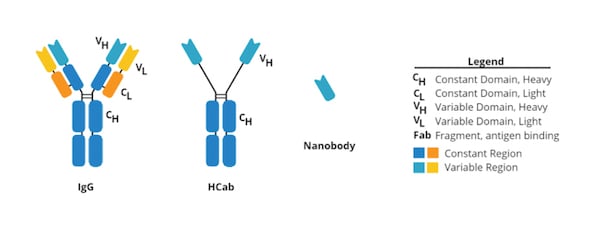 |
| Figure 1: Comparison of common antibody, HCab, and nanobody. For more information read our secondary nanobodies blog post. |
A rapid selection platform for generating sybodies against membrane proteins
Developing antibodies against proteins embedded in membranes is tricky because they have fewer hydrophilic surfaces, require detergents or lipids to remain folded, and are often less abundant than some other proteins. The Seeger lab developed a robust in-vitro protocol that enables rapid selection of synthetic nanobodies, called sybodies, against membrane protein targets in as little as 3 weeks (Zimmermann et al., 2020). This selection platform was designed so that any standard lab can quickly select for sybodies targeting a membrane protein of interest using the Sybody Generation Toolbox and commercially available reagents in three major steps - all without the need to find a llama farm!
To find an optimally binding sybody, you want to start out with a large and diverse pool of sybodies that hypothetically contain ones that bind strongly to your target protein. The Seeger lab designed 3 different mRNA sybody libraries for use in the protocol, which cover 3 naturally-occurring nanobody structures (concave, loop, and convex). Each of these 3 mRNA libraries have a concave, loop, or convex scaffold and are randomized with a defined set of different trinucleotides (corresponding to different amino acids) in the regions that bind to the antigen. The Seeger lab can directly provide the three sybody libraries in the form of mRNA for academic research.
The template plasmids for generating your own libraries are also available in the Sybody Generation Toolbox plasmid kit: SB_concave, SB_loop, SB_convex. These scaffolds contain serines and threonines at positions that can be randomized for creating the libraries (Zimmermann et al., 2018).
Find the Sybody Generation Toolbox!
Starting with libraries and a biotinylated (for purification purposes) protein target of interest, the protocol goes through 3 major steps to select for optimal sybody candidates.
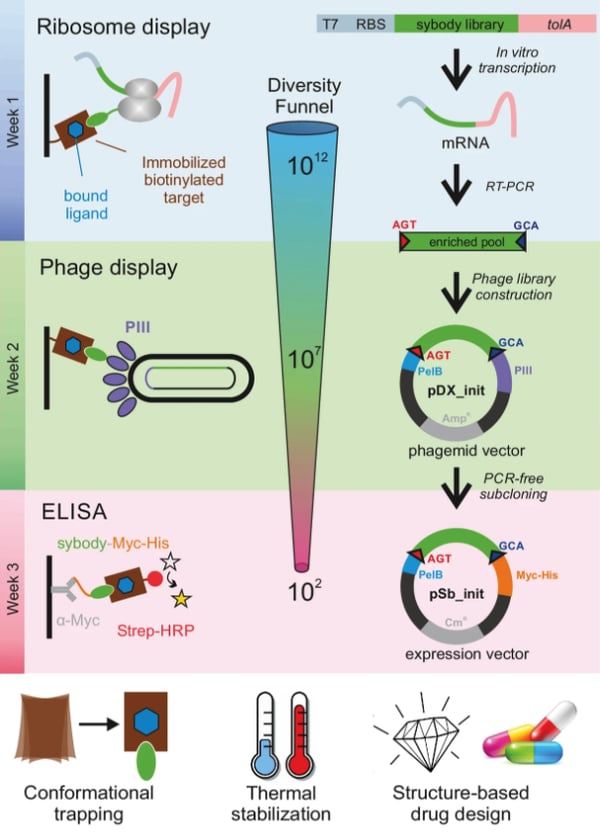 |
| Figure 2: Outline of the process for selection of sybodies against membrane proteins. Image from Zimmermann et al., 2018. |
Ribosome display
The Seeger lab starts with a “pre-enrichment” selection step using ribosome display because it has the advantage of starting out with a larger initial starting library size (up to 1012 members) in comparison to other display systems using phage and yeast (109-1010 members). This selection step involves using a commercial kit for in-vitro translation of the mRNA libraries. Each mRNA molecule in the library doesn’t encode a stop codon, which causes the ribosome to pause at the end of the mRNA molecule, forming a ribosome complex consisting of the translated protein (sybody), ribosome, and mRNA. During the screening process, displayed sybodies that bind to the biotinylated target protein can be recovered for the subsequent steps.
Phage display
mRNA from the enriched pool of sybody candidates are then reverse transcribed into cDNA fragments that are efficiently subcloned into the phagemid pDX_init plasmid, using the fragment exchange (FX) cloning technique which uses Type IIS restriction sites (Geertsma, 2013). Once the phagemids are constructed, they are used to produce a phage library consisting of phages that display the candidate sybodies in their coat protein to screen for binding to the target protein. The protocol describes going through two rounds of phage display to increase the enrichment of target-specific sybodies.
Final enrichment via enzyme-linked immunosorbent assay (ELISA)
The enriched sybody pools from the phage display screens are subcloned (using FX cloning) into the expression vector pSb-Init, which attaches a Myc-tag and a His6-tag to the end of each sybody candidate. These tags are used in a final enrichment step for identifying sybodies that bind strongly and specifically to the protein of interest. The Seeger lab used the ELISA technique described in their protocol because it is less susceptible to identifying unspecific and low-affinity sybodies (Zimmermann et al., 2020). This step of the protocol starts out with anti-Myc antibodies bound to an ELISA plate to capture Myc-tagged sybodies. Then, the biotinylated target protein is added and after a few washes, binding is detected using a colorimetric reaction.
After the ELISA step, the narrowed-down pool (usually 10-30) of sybody candidates are sequenced for identification. All unique sybodies can be subcloned into pBXNPH3 or pBXNPHM3 expression plasmids for the production of tag-free proteins for further characterization and analysis.
In a rapid response to the COVID-19 pandemic, the Seeger lab demonstrated they could use the protocol to rapidly identify and isolate sybodies targeting the SARS-CoV-2 Receptor Binding Domain found on the membrane-bound spike protein, in a remarkable 12 working days. While the lab used this method for COVID-19 research, this protocol holds promise for developing treatments for other diseases or for studying other membrane-bound proteins.
The full protocol for this sybody selection platform can be found here and relevant plasmids are available in the Sybody Generation Toolbox kit.
Find the Seeger lab's SARS-CoV-2 sybodies!
References
- Geertsma ER (2013) FX Cloning: A Versatile High-Throughput Cloning System for Characterization of Enzyme Variants. In: Methods in Molecular Biology. Humana Press, pp 133–148. https://doi.org/10.1007/978-1-62703-293-3_10
- Walter J et al. (2020) Synthetic nanobodies targeting the SARS-CoV-2 receptor-binding domain. BioRxiv. https://doi.org/10.1101/2020.04.16.045419.
- Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, Hoffmann M, Pöhlmann S, Graham BS, Callewaert N, Schepens B, Saelens X, McLellan JS (2020) Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 181:1004–1015.e15 . https://doi.org/10.1016/j.cell.2020.04.031
- Zimmermann I, Egloff P, Hutter CAJ, Kuhn BT, Bräuer P, Newstead S, Dawson RJP, Geertsma ER, Seeger MA (2020) Generation of synthetic nanobodies against delicate proteins. Nature Protocols 15:1707–1741. https://doi.org/10.1038/s41596-020-0304-x
- Zimmermann I, Egloff P, Hutter CA, Arnold FM, Stohler P, Bocquet N, Hug MN, Huber S, Siegrist M, Hetemann L, Gera J, Gmür S, Spies P, Gygax D, Geertsma ER, Dawson RJ, Seeger MA (2018) Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7: . https://doi.org/10.7554/elife.34317
Additional resources on the Addgene blog
- Opto-Nanobodies: Using Light to Manipulate Cell Signaling and Protein Purification
- RANbodies: Reporter Nanobody Fusions
- Plasmids 101: Secondary Nanobody Toolbox
Resources on Addgene.org
- View the Antibody plasmid collection
- Browse the RESOLUTE solute carriers collection
- Find other COVID-19 and Coronavirus Plasmids & Resources
Topics: Other Plasmid Tools, Plasmids, COVID-19, Antibodies






Leave a Comment