 One of the best things about sharing plasmids through Addgene is that we provide an added level of confidence in the plasmids we distribute through our quality control processes. Every plasmid we receive is rigorously verified before becoming available to the community.
One of the best things about sharing plasmids through Addgene is that we provide an added level of confidence in the plasmids we distribute through our quality control processes. Every plasmid we receive is rigorously verified before becoming available to the community.
This is no small task, however, at a repository that consistently receives around 200 new DNA samples every week. Here we will provide an inside look at the steps we take to verify the identity and quality of the plasmids we make available and provide some advice on the steps you can take to verify your own plasmids.
Staying organized
With hundreds of samples coming through our doors, the first step to ensuring plasmid quality is to stay organized. Here at Addgene, we collect data on every plasmid we distribute through our deposit process, and then we carefully track samples through our lab using barcoded tubes and plates.
The critical pieces of information for a given plasmid are the backbone, inserted gene/functional mutations, antibiotic resistance, restriction sites used during cloning, and most importantly, the predicted full plasmid sequence. Other information may also be critical to the maintenance and use of certain plasmids, such as special growth instructions (temperature, strain, media supplements), suggested sequencing primers, or mammalian selection markers.
You don’t need a fancy database to organize your own plasmid collection, although you will want to keep digital copies of your predicted sequence information. Collect plasmid information in a binder or digital notebook and make sure you note the freezer location of every plasmid in your collection. Use one of the many freely-available DNA-editing software packages to assemble your expected full plasmid sequences (examples include Benchling, Serial Cloner, and Snapgene), and save these to a shared computer or server. This will ensure that future members of your lab can access and use the materials they need, and avoid duplicating your effort. In addition, if you should choose to deposit your plasmids with us, it will be easy to find all of the information needed for the process.
Always sequence verify
Once the physical sample and its associated information are at Addgene, we are ready to sequence verify the plasmids. This is the most important step in ensuring quality, as it will catch any errors made during the cloning, sample handling, or sequence assembly steps.
All of our incoming plasmids are sequenced at least twice, and sometimes more depending on the number of features we need to verify.
Determining which features to verify will depend on the particular plasmid you are working with, but, in most cases at Addgene, we check what we call the “insert,” (the gene or genetic element cloned into the plasmid) and any other important features that differentiate this plasmid from its predecessor. Important features may include cloning junctions (where most sequence assembly errors occur), deletions, mutations, in-frame protein fusions, and multiple cloning sites. In the simplest case we have a standard backbone with one insert, and can use common primers to sequence from the backbone into the insert. If the insert is large, we may only check the 5’ and 3’ ends to verify that the sequence assembly and identity of the insert are correct. In other cases, we deal with multiple inserts or internal mutations that require more than two sequencing reactions.
In the more complex cases, we sometimes have to design new primers to sequence the important features. You can use primer design software such as Primer3 to help you design your custom primers. Remember, when designing a sequencing primer, you need some space between your primer binding site and the first feature you want to verify because you won’t get reliable sequence data back for the first 50-100 base pairs of your sequencing read. You can expect to recover around 750 bp of clean sequence from a typical reaction, or about 250 amino acids worth, so make sure the feature you want to verify is within this range.
Read some of our other blog posts for more tips on sequence analysis, plasmid verification, and using NCBI BLAST.
Tips for high-throughput plasmid sequencing
If you’re like us, you need a strategy for processing several sequencing reactions in parallel. We have a few tricks to simplify the process.
First, nearly all of our Sanger sequencing is done from glycerol stocks (overnight cultures with 15% glycerol), as opposed to purified DNA or colony sequencing. This means that we are sequencing from the exact same stock that we store and distribute from. Of course, your culture should always be derived from a single colony to ensure uniformity of the culture. Sequencing directly from the glycerol stock saves us the step of purifying the DNA and gives comparable success rates and sequence quality, which is critical for the efficient handling of large numbers of samples.
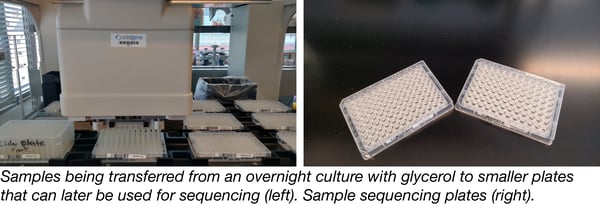
When we grow overnight cultures for glycerol stock production and storage, we grow a little extra and transfer 100 µl of glycerol stock to each of 2 96-well microplates (identical copies, see image above). We then select forward and reverse primers for each sample with the help of DNA-editing software which integrates our database of over 2000 primers. Two 96-well PCR plates are prepared with the primer stocks for forward and reverse reactions, respectively, and then shipped along with their corresponding glycerol plate on dry ice to the sequencing provider.
Each sequencing service will have different requirements for volume and concentration, and not all providers are equipped to perform Sanger sequencing from glycerol stocks, so make sure to select the appropriate service and read the submission instructions.
As for the analysis of a large number of sequencing reactions, you can make your life much easier by selecting the right primer. This often starts by determining what features you expect to differ between plasmids, and selecting or designing a primer specifically to target this region, as outlined above. Avoid primers that bind within or must read through repetitive regions (such as LTRs or ITRs), hairpins, or strong terminators. Detailed instructions on other factors to consider when designing your primers can be found here.
Finally, you will want to have a well-organized database of expected sequences, either for the inserts or the full plasmids. This will enable you to quickly BLAST your sequencing result against the expected sequence for quick verification, which can be done through the align tab of our sequence analysis web-tool. Alternatively, you can use DNA-editing software to perform alignments of multiple sequences to visually check for homology and mismatches.
Do you have your own best sequencing practices or maybe some tips on how to get better sequencing reads? Tell us your favorite sequencing tricks in the comments section below.
Additional Resources on the Addgene Blog
- Get 6 Tips for Analyzing and Troubleshooting DNA Sequence
- Learn Additional Ways to Verify Your Plasmid
- Learn How Addgene Uses Barcodes to Track Samples
Resources on the Addgene Website
- View our Plasmid Reference Pages
- Get Plasmid Backbone Information from Our Vector Database
Topics: Other Plasmid Tools, Plasmids






Leave a Comment