Many neuroscience experiments that require gene expression in a specific cell type rely on transgenic models that express recombinases like Cre or Flp in their cells of interest and recombinase-dependent AAV vectors for selective transgene expression. While this is a powerful experimental strategy, one of the most common concerns we get here at Addgene about this system is off-target or spurious expression of recombinase-dependent AAV vectors. In this article, we will discuss some of the causes of off-target expression and ways to optimize experiments to minimize it.
Overview of DIO/FLEx vector design
Recombinase-dependent vectors utilize pairs of specific sequences, lox (recognized by Cre) or frt (recognized by Flp), to either flip or excise a gene that is located between them. Early recombinase-dependent vector designs used a lox-STOP-lox (LSL) or frt-STOP-frt (FSF) sequence prior to an ATG start codon to prevent transcription in cells where the recombinase was not present (Kuhlman, SK. 2008). While theoretically this strategy should work, these vectors suffered from high levels of spurious expression.
As such, researchers developed new tools called FLEx/DIO vectors where the transgene is floxed by two pairs of recombination sites and the open reading frame is inverted in relation to the promoter, hence the name, double-floxed inverted open-reading frame (DIO), or FLip-EXcision (FLEx) vector. The two recombination sites are both recognized by Cre, but they are comprised of different sequences (loxp and lox2272) and can not recombine with each other. When Cre is introduced to the system it binds one pair of lox sites, let's say the loxp pair, and flips the transgene into the correct orientation for transcription. Upon recombination one of lox2272 sites will now be sandwiched between 2 loxp sites and is excised by Cre (Figure 1). This results in one loxp site and one lox2272 site on either end of the transgene. Since loxp and lox2272 can not recombine, this ensures that the transgene is no longer able to flip back into the inverse orientation (See Plasmids 101: FLEx Vectors). While this strategy greatly reduced the overall level of off-target transgene expression, low levels of spurious expression were still observed in many systems. So what is the cause of this spurious expression?
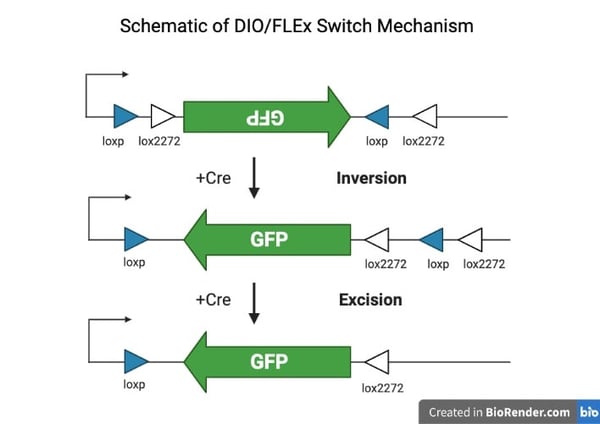 |
| Figure 1. Cre-recombinase binds to either pair of lox sites, let's say the lox2272, that are in the opposite orientation (direction of arrows) the inverted transgene is reverted into the sense orientation and one of the unused (loxp) sites will be flanked by two lox sites now in the same orientation. Next, the loxp site is excised since it is between two lox2272 sites in the same orientation preventing the transgene from reverting. Figure created with BioRender.com |
Causes of recombination and leaky expression
While leaky expression and spontaneous recombination is a known problem of DIO/FLEx vectors, the cause of these events, until recently, was not known. A recent paper from Ed Callaway’s lab (Fischer, KB. 2019) set-out to systematically test the potential causes of these phenomena by analyzing the plasmid and viral DNA sequences and expression profile at multiple steps from plasmid amplification in bacteria to transgene expression in vivo.
Since DIO/FLEx vectors contain two sets of homologous recombination sites, they hypothesized that spontaneous recombination between these sites may be the cause of leaky expression. To test their hypothesis, they randomly scrambled one pair of lox sites and either used the same sequence to flank a transgene (homologous pairs) or used different scrambled sequences to flank the transgenes (non-homologous pairs) (Figure 2.). They found that almost all off-target expression can be attributed to spontaneous recombination between homologous pairs, and that the degree of homology is correlated with the level of spontaneous recombination. This indicates that the amount of leaky expression can largely be attributed to the level of spontaneous recombination between homologous pairs before viral packaging even begins. This spontaneous recombination persists even in recombinase deficient E. coli and under conditions used to minimize recombination that are typically employed during viral plasmid amplification (Fischer, KB. 2019). This means that all DIO/FLEx AAV vectors will have some level of recombination that can lead to leaky expression in cells that do not express Cre or Flp.
Here at Addgene we quantify the amount of transgene recombination for every lot of AAV vector we produce containing a DIO/FLEx cassette using next-generation sequencing (NGS) (Guerin, K. 2020). In general, we observe that 0.1 - 0.8% of packaged viral genomes contain a recombined/functional transgene that can be expressed in cells where Cre is absent. Additionally, we have experienced that some vectors are much more prone to recombination and can exhibit recombination levels as high as 1-3%. In light of this finding, we now include guidance on the plasmid page of DIO/FLEx AAV vectors where this occurs to alert the scientist to exercise caution when using them.
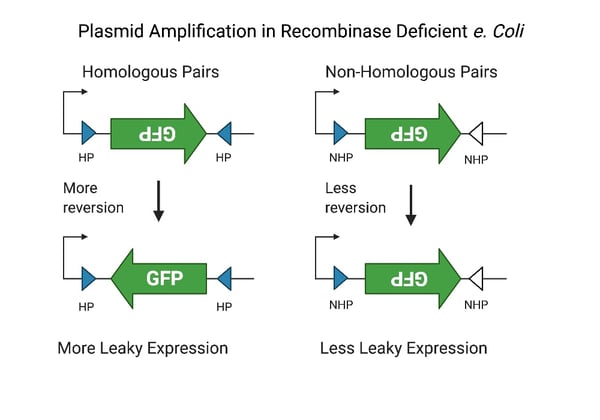 |
| Figure 2. Schematic diagram showing the mechanism of spontaneous reversion during AAV plasmid amplification. On the left homologous pairs of scrambled cre-recombinase sites lead to greater frequency of reversion and leaky expression. On the right non-homologous scrambled recombination sights lead to reduced reversion and less off-target expression. Figure created with BioRender.com |
Strategies for optimizing expression
So now that we know spontaneous recombination occurs with AAV DIO/Flex vectors and leaky expression can occur in cells that don’t express Cre or Flp, what can we do about it?
Test the serotype and promoter
The first step is to know how the AAV vector you intend to use performs in your system. This includes a thorough testing of both the serotype and promoter with in vivo experiments to determine the optimal/minimal injection titer and volume needed to achieve adequate expression in your cells of interest.
The AAV serotype used can exhibit profound differences in tropism between cell types (astrocytes, microglia, neurons, etc.) and even neuronal phenotype in a mixed population (Burger, C. 2004, Aschauer, DF. 2013, O’Carroll, SJ. 2021). This is due to differences in the expression of receptors, post-translational modifications, and other factors that AAV utilizes to gain entry into the cell (Dudek, AM. 2018).
If the serotype you are using transduces cells that you are not interested in with greater efficiency, it could lead to greater off-target expression just due to more overall viral particles getting into those cells. Therefore, the first step in any AAV experiment is to understand which serotype best transduces your cells of interest. We recommend testing several different serotypes to determine which one is the best for your cells of interest.
Determine injection titers and volumes based on the serotype
Since each serotype also exhibits distinct patterns of spread from the injection site it is advisable to try a few different injection titers and volumes to determine the optimal injection parameters needed to transduce your cells of interest. Using a constitutively expressing vector for this step may save time as it reduces variability that may be encountered with DIO/FLEx vectors and off-target expression making it easier to determine the spread pattern around the injection site.
Test the promoter
It is also essential that the promoter be thoroughly tested in your cells. Some promoters, such as CAG are very strong and may require less viral particles or relatively short expression times, where as others, such as hSynapsin, are relatively weak and may require longer waiting periods and/or higher injection titers to achieve adequate expression. Conducting a time course experiment to check expression levels over several weeks allows the researcher to determine when the optimal time to conduct an experiment begins. This can save you time in the long run since not having this information could lead to waiting longer than necessary before starting your experiment, or if you start an experiment before adequate expression is reached you may obtain a false negative or conclude the viral vector is not working. When using a “cell-type specific” promoter, such as the truncated tyrosine hydroxylase (TH) promoter for catecholamine neurons or glial fibrillary acidic protein (GFAP) for astrocytes it is important to test it in the brain region of interest, especially when using DIO/FLEx vectors for off-target expression. Some classes of neurons transiently express TH during development or have low levels of TH mRNA without expressing TH protein indicating that there is promoter activity. Additionally, some neurons express low levels of GFAP, which can lead to off-target expression of the packaged transgene in neurons and glia.
Check the recombination rate
The first step in optimizing DIO/FLEx vectors is to know the recombination rate of the vector you are using. Contact Addgene (help@addgene.org), or the supplier of your vector, to obtain this information.
Optimize titer for the DIO/FLEx vector
Once you have determined the optimal injection parameters for the serotype that best transduces your cells of interest, you can now begin to optimize the DIO/FLEx vector. This step is essentially the same as what was used to optimize the serotype, except now you will already have a starting “optimal” injection volume and titer to begin with. Now you will mainly only have to adjust the titer in order to minimize off-target expression while still achieving adequate expression in your cells of interest. You should now be able to make smaller adjustments to a set of injections into your animal model. For example, in a recent study by the Wickersham lab (Lavin, TK. 2020) it was determined that the optimal titer to prevent off-target expression of the sensitive FLEx-split TVA receptor used for tracing neural pathways was 8.5 x 10e10 GC/mL. The lot of viruses used (100798-AAV, lot #v15287, titer 1.7x10e13 GC/mL and 100799-AAV1, lot # v14715, titer 2.4x10e13 GC/mL) for this study were packaged at Addgene and had a relatively low recombination rate (0.1% and 0.13% respectively) still required a 200-300 fold dilution to minimize off-target expression. Since recombination rates vary between every lot of AAV vector, a range of dilutions and/or injection volumes may need to be tested with each new vector or lot used.
Following this stepwise optimization will take a fair amount of time, and it may be tempting to skip some steps and get to experiments. However, by optimizing all conditions first, you will save time, frustration, and funds in the long-run by ensuring you are minimizing any off-target expression that can confound your results.
Conclusion
Using AAV vectors in your experiments requires a full understanding of vector performance in your system. It is very rare for a methods section to include all of the variables that need to be accounted for (i.e. serotype, promoter, titer, buffer, etc.) and this can lead to the misconception that if one uses a vector with the same titer and serotype previously used in a different brain region that the same result will be achieved. The fact is that there are many known and unknown factors that affect vector performance in each and every system and cell type. Therefore, it is essential that every new vector, whether it be a completely new transgene or merely a new lot, be thoroughly tested in the researcher's system before beginning large scale experiments. This will help to ensure the accuracy and reproducibility of the data and can instill greater confidence when interpreting the outcome.
References and resources
References
Aschauer DF, Kreuz S, Rumpel S (2013) Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain. PLoS ONE 8:e76310. https://doi.org/10.1371/journal.pone.0076310
Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N (2004) Recombinant AAV Viral Vectors Pseudotyped with Viral Capsids from Serotypes 1, 2, and 5 Display Differential Efficiency and Cell Tropism after Delivery to Different Regions of the Central Nervous System. Molecular Therapy 10:302–317. https://doi.org/10.1016/j.ymthe.2004.05.024
Dudek AM, Pillay S, Puschnik AS, Nagamine CM, Cheng F, Qiu J, Carette JE, Vandenberghe LH (2018) An Alternate Route for Adeno-associated Virus (AAV) Entry Independent of AAV Receptor. J Virol 92. https://doi.org/10.1128/jvi.02213-17
Fischer KB, Collins HK, Callaway EM (2019) Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc Natl Acad Sci USA 116:27001–27010. https://doi.org/10.1073/pnas.1915974116
Guerin K, Rego M, Bourges D, Ersing I, Haery L, Harten DeMaio K, Sanders E, Tasissa M, Kostman M, Tillgren M, Makana Hanley L, Mueller I, Mitsopoulos A, Fan M (2020) A Novel Next-Generation Sequencing and Analysis Platform to Assess the Identity of Recombinant Adeno-Associated Viral Preparations from Viral DNA Extracts. Human Gene Therapy 31:664–678. https://doi.org/10.1089/hum.2019.277
Lavin TK, Jin L, Lea NE, Wickersham IR (2020) Monosynaptic Tracing Success Depends Critically on Helper Virus Concentrations. Front Synaptic Neurosci 12. https://doi.org/10.3389/fnsyn.2020.00006
O’Carroll SJ, Cook WH, Young D (2021) AAV Targeting of Glial Cell Types in the Central and Peripheral Nervous System and Relevance to Human Gene Therapy. Front Mol Neurosci 13. https://doi.org/10.3389/fnmol.2020.618020
Additional resources on the Addgene blog
- Plasmids 101: FLEx Vectors
- Important Considerations When Using AAVs
- Three Key Considerations For Precise Neuronal Targeting Using AAV Technologies
Resources on Addgene.org
- Find ready to use AAV vectors at Addgene.
- Find AAV plasmids to use in your experiments.
Topics: Viral Vectors, Viral Vector Protocols and Tips, AAV





Leave a Comment