🎶 “Bye bye L-arabinose drive
Put your cultures in the shaker
Turn the LEDs on
And when you want
You can just turn them off
No need for any wash
No need for any wash…” 🎶
- Barbara Di Ventura
If you follow Barbara Di Ventura on Twitter, you might have seen the video of her singing about the paper recently published by her lab and Mustafa Khammash's lab at ETH Zürich. “Instead of a graphical abstract, it’s a musical abstract,” says Di Ventura, a professor at the University of Freiburg. This paper is about a new tool called BLADE (blue light-inducible AraC dimers in E. coli) that turns expression on and off with blue light.
Inspired by @UriAlonWeizmann, here my song about our paper recently published in @nchembio
— Barbara Di Ventura (@barbara_synbio) May 13, 2021
Title: Bye bye L-arabinose drive. Dedicated to Edo, Armin and @KhammashLab. I hope you all enjoy it!
Disclaimer: I had to cut it very abruptly to stay within the time limit. pic.twitter.com/j6rhCCby2m
The original AraC transcriptional regulator
Their system is based on the AraC protein that controls transcription from the PBAD promoter. “AraC is widely used in synthetic biology, biotechnology, and almost every microbiology lab,” says Di Ventura. It relies on the sugar arabinose to activate transcription by causing conformation changes in the AraC dimer. In doing so, the dimer shifts from one that represses transcription to one that activates transcription.However, this system lacks spatio-temporal regulation, and is difficult to reverse, making it unsuitable for some types of experiments.
(To get more information on AraC and other promoter systems, head over to our Plasmids 101: Inducible Promoters article.)
Modifying AraC to create BLADE
The team modified AraC to create BLADE by replacing its dimerization domain with VVD, a light-responsive domain that dimerizes upon exposure to blue light (Fig. 1). Another blue light-inducible dimerization domain, Vaucheria frigida aureochrome1 (VfAu1), led to functional BLADE variants, too. The team tested different linkers between VVD and the DNA-binding domain of AraC using a mCherry reporter downstream of a PBAD promoter. While none of the constructs gave as strong a signal as with the wild-type AraC, it still reached the level of expression needed for most applications.
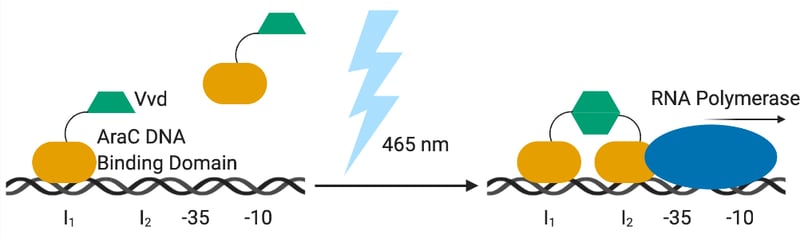 |
| Figure 1: With blue light, the VVD domains of the BLADE dimerize and activates transcription. |
The team created two constructs, among others:
- pBLADE-mCherry: This construct, built from the pBAD33 backbone, contains BLADE, the PBAD promoter, and a mCherry reporter downstream of the PBAD promoter. To use this plasmid with your gene of interest, you would replace mCherry with your gene of interest.
- pBLADEONLY: This plasmid contains only BLADE and is based on the pTRC99a plasmid. It does not include the PBAD promoter. If you already have a pBAD33-based plasmid expressing your gene of interest, all you have to do is co-transform pBLADEONLY. That way, a previously cloned gene of interest, under the pBAD promoter, can be controlled with light without the need of any cloning.
What advantages does BLADE have over AraC?
When compared to the original AraC system, BLADE is simply more precise. “We show that you have less heterogeneity in the population and you can do all of these more spatially confined experiments as well as those requiring reversibility that you cannot do with AraC,” says Di Ventura.
BLADE activates transcription with the flick of a light switch and shuts off just as quickly. The original AraC system, which uses arabinose, requires many wash steps to remove arabinose making it difficult to regulate quickly.
Arabinose also circulates within growth media and can’t be contained to a specific area. Blue light, however, is spatially confined and you can see this demonstrated through living images created by the team (Fig. 2). These images are based on the concept of bacteriographs created about a decade ago. To create these images, the scientists used a strain transformed with pBLADE expressing superfolder GFP instead of mCherry under the PBAD promoter. They created a bacterial lawn on an agar plate, put a mask over the plate containing the pattern of the image, and shined blue light over the plate. Areas on the plate that are blocked by the mask will not produce the fluorescent protein while areas that are exposed will fluoresce. With these images, you can see just how spatially precise the activation is.
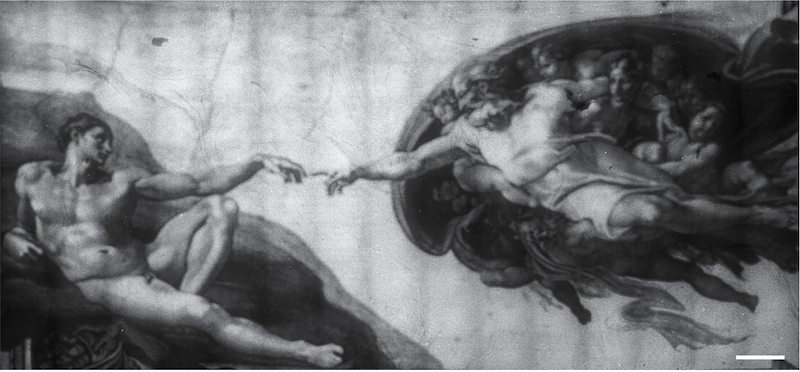 |
| Figure 2: A bacteriograph of Michelangelo's "The Creation of Adam." This bacteriograph was created using 160 individual images stitched together. Image from Romano et al., 2021 with permission. |
BLADE is just one of the several optogenetic tools that Di Ventura’s lab and Khammash's lab have created and shared with Addgene. You can find all the plasmids deposited from Di Ventura's lab, including the popular light-inducible nuclear export system (LEXY) here. Optogenetic plasmids from the Khammash lab can also be found here.
References and Resources
References
Romano E, Baumschlager A, Akmeriç EB, Palanisamy N, Houmani M, Schmidt G, Öztürk MA, Ernst L, Khammash M, Di Ventura B (2021) Engineering AraC to make it responsive to light instead of arabinose. Nat Chem Biol. https://doi.org/10.1038/s41589-021-00787-6
Additional resources on the Addgene blog
- Learn more about inducible promoters
- Visit the Plasmids topic overview page
Resources on Addgene.org
- Check out the Molecular Biology Reference
- Download Addgene's Plasmids 101 eBook
Topics: Optogenetics, Microbiology, Expression Vectors, Plasmids






Leave a Comment