 This post was contributed by Tim Herman, director of the MSOE center for BioMolecular Modeling and the CEO of 3-D Molecular Designs.
This post was contributed by Tim Herman, director of the MSOE center for BioMolecular Modeling and the CEO of 3-D Molecular Designs.
Have you ever held your favorite protein in the palm of our hand? Well, actually – have you ever held a model of your favorite protein in your hand? At 3D Molecular Designs, we create physical models of proteins using 3D printing technology. The tagline for our company is “…where molecules become real”. We sometimes forget that the models we create are around 30 million times larger than the real thing. Nevertheless, the models are compelling, and I encourage you to hold one.
Why do we need physical models of protein structure?
Structural biologists have always struggled to effectively communicate the nuances of protein structure in a flat 2-D images. In the early days of structural biology, Byron Rubin, a crystallographer working with Jane Richardson, provided an initial solution to this problem with the development of Byron’s Bender – a mechanical device that could be used to bend 1/8th inch steel wire into a backbone structure of a protein, based on its atomic coordinates.
These wire models were useful in that they not only traced the 3-D path of the protein’s backbone in three-dimensional space, but their flexible and jiggly nature suggested the dynamic nature of a protein. The Bender’s popularity was short-lived as powerful molecular graphics packages were soon developed that allowed researchers to create stunning, interactive images of their proteins.
Since the 1980’s, journal articles have been filled with impressive, full-color images of proteins. But they are still flat. To truly appreciate the 3D structure of these proteins, it is necessary to download a pdb file of the structure’s atomic coordinates, and “open” these structures with molecular viewing software. In reality, this rarely happens. But if you ever have an opportunity to read one of these structure papers – while holding a physical model of the protein in your hand – you will understand the power of a physical model. Until then, you can only wonder what I am talking about.
How do researchers use physical models of protein structure?
We have been creating physical models of proteins for researchers for the past fifteen years. On those occasions when we are fortunate to hand-deliver the model, the following sequence of events plays itself out: The researcher breaks eye-contact with any other human in the room. Their eyes are focused on the model as they turn it over and over in their hands. They then start talking to themselves as they compare the features displayed on the model with their mental model of the protein. At some point during this interrogation process, they will almost invariably say something like “Oh,…I didn’t quite appreciate that….”
For example, when Tom Steitz was presented with a physical model of his large ribosomal subunit, he kept sliding his palm across the surface of the protein where the nascent polypeptide emerges from the exit channel. He then said “I hadn’t realized how perfectly flat the complex is on this surface”….where the complex is docked on the membrane of the endoplasmic reticulum. And graduate students, momentarily forgetting that the physical model is based on their own modeling of the protein’s structure into experimentally determined electron density, have been known to say something silly like “See there, arginine 53 really is right there, at the end of helix 3…just like we said it was”. Seeing is believing. And in that sense, models can be both useful, and dangerous.
Subscribe to CRISPR Blog Posts
How do educators and students use molecular models?
We created the Center for BioMolecular Modeling (CBM) in 1999 with the expressed purpose of creating physical models of proteins and other molecular structures for use in professional development programs for bioscience educators. Using these hands-on tools, educators introduce their students to this otherwise invisible molecular world. For the students, the models force them to slow down and focus their attention on something tangible. Under the guidance of a trained educator, these instances of focused attention become magical teaching moments. The models function as thinking tools – eliciting questions like “what does this model representt?" ... "how does this protein function in the human body?” … and probably most importantly… "how do we know this?”
Over the past fifteen years, the CBM has developed a student-centered SMART Team modeling program in which small teams of students work closely with local researchers to learn about their research while creating physical models of proteins that are being investigated in their labs.
Soon after we began working with educators in our Center for BioMolecular Modeling, we spun off a small, family-run business – 3D Molecular Designs – to commercialize some of the best instructional materials that resulted from our work with teachers, and to create custom models of proteins for researchers.
Modeling CRISPR
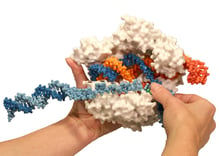
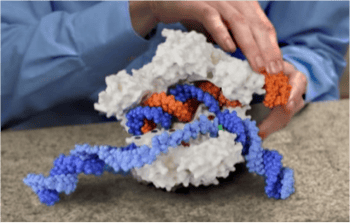 We were excited when Jacob Corn contacted us in 2014 to ask if we could make a model of Cas9 based on the structure published by Martin Jinek’s group. We had already made an initial model of this protein – but were very confused because the HNH nuclease active site wasn’t even close to its reported cleavage site on the complementary DNA strand. And this is why we love working with researchers on a model. The little knowledge that we have when we jump into a new modeling project can be a dangerous thing. Fortunately, Jacob was able to quickly explain that it was common knowledge within the CRISPR community that this first Jinek structure had caught the protein in a conformation that was not “poised for cleavage”. Jacob computationally added the non-template strand to the Jinek structure so that the resulting physical model is an effective teaching tool. You can read about Jacob’s initial reaction to this model on his blog. Or, for a description of how effective this model was in explaining CRISPR technology to 4th graders, check out Jennifer Doudna’s new book A Crack in Creation.
We were excited when Jacob Corn contacted us in 2014 to ask if we could make a model of Cas9 based on the structure published by Martin Jinek’s group. We had already made an initial model of this protein – but were very confused because the HNH nuclease active site wasn’t even close to its reported cleavage site on the complementary DNA strand. And this is why we love working with researchers on a model. The little knowledge that we have when we jump into a new modeling project can be a dangerous thing. Fortunately, Jacob was able to quickly explain that it was common knowledge within the CRISPR community that this first Jinek structure had caught the protein in a conformation that was not “poised for cleavage”. Jacob computationally added the non-template strand to the Jinek structure so that the resulting physical model is an effective teaching tool. You can read about Jacob’s initial reaction to this model on his blog. Or, for a description of how effective this model was in explaining CRISPR technology to 4th graders, check out Jennifer Doudna’s new book A Crack in Creation.
Making models more useful through flexibility
 It is common knowledge in the education community that All models are wrong….Some models are useful. While a physical model of a protein might represent one aspect of the protein very well, it will almost certainly fail to convey other aspects of the structure. While this first Cas9 model is certainly useful, it is also wrong in at least one serious way. We represented the Cas9 protein in a surface format made of rock-hard, inflexible plaster. As a result, the model obscures the flexible nature of all proteins in general, and the amazing conformational flexibility that the Cas9 protein must have in order to scan along the double-stranded DNA, recognize a PAM sequence, allow the complementary base pairing between the guide RNA and the target DNA strand, and then activate two different endonuclease active sites.
It is common knowledge in the education community that All models are wrong….Some models are useful. While a physical model of a protein might represent one aspect of the protein very well, it will almost certainly fail to convey other aspects of the structure. While this first Cas9 model is certainly useful, it is also wrong in at least one serious way. We represented the Cas9 protein in a surface format made of rock-hard, inflexible plaster. As a result, the model obscures the flexible nature of all proteins in general, and the amazing conformational flexibility that the Cas9 protein must have in order to scan along the double-stranded DNA, recognize a PAM sequence, allow the complementary base pairing between the guide RNA and the target DNA strand, and then activate two different endonuclease active sites.
Fortunately, there is never just one model of a protein. We now have another model of Cas9 – a flexible alpha-carbon backbone model made of nylon – that more accurately suggests the potential of this protein to undergo conformational shifts. This backbone model of Cas9 is based on Fuguo Jiang’s structure of Cas9 – primed for cleavage. This structure captures the displaced non-target DNA strand and shows the HNH nuclease domain swung around in close juxtaposition to its cleavage site on the target DNA strand. In addition, this model also displays the several Lys/Ala mutations reported by Ian Slaymaker and colleagues to reduce off-target hits in a rationally engineered Cas9 with improved specificity. Hold this flexible model in your hand for a while and you will wake up in the middle of the night dreaming about the conformational gymnastics that this protein must undergo as it interrogates your genome.
If you are interested in experiencing Cas9 up close and personal, visit the 3D Molecular Designs web site. Or, if you are visiting a local classroom or trying to explain your science to the general public, visit the Center for BioMolecular Modeling web site to check out our hands-on teaching tools that are designed to make the molecular world real for students.
Many thanks to our guest blogger Tim Herman!
 TIm Herman is the director of the MSOE center for BioMolecular Modeling and the CEO of 3-D Molecular Designs.
TIm Herman is the director of the MSOE center for BioMolecular Modeling and the CEO of 3-D Molecular Designs.
Additional Resources on the Addgene Blog
- Learn All about CRISPR
- Use CRISPR for Genome Visualization
- Tips for a First Time CRISPR User by a First Time CRISPR User
Resources on Addgene.org
- Visit Our CRISPR Guide Page
- CRISPR/Cas9 History
- Browse All CRISPR Plasmids and Resources
Topics: Scientific Sharing, Education







Leave a Comment