This post was contributed by Katherine Rogers, a postdoctoral researcher at the Friedrich Miescher Lab of the Max Planck Society.
 Zebrafish (Danio rerio) have been used since the 1930’s in a range of biological studies, including investigations into environmental pollutants and health, embryo growth, brain function, and disease development. Why have zebrafish become such a popular model organism?
Zebrafish (Danio rerio) have been used since the 1930’s in a range of biological studies, including investigations into environmental pollutants and health, embryo growth, brain function, and disease development. Why have zebrafish become such a popular model organism?
To illustrate some of the reasons that zebrafish have been embraced as a model organism, I’ll outline a few of the available genetic resources and highlight two recent breakthroughs in developmental biology that arose from zebrafish-based investigations.
Genetic resources for zebrafish
A common approach to unravel the gene networks that drive biology is to examine the consequences of manipulating genes. One reason for the success of zebrafish as a model organism is its amenability to genetic manipulation. Tools for gene manipulation together with information about the genome are powerful resources for investigating any biological process.
High quality genome sequence
A high-quality sequence assembly of the zebrafish genome was published in 2013, revealing that about 70% of human genes have a counterpart in zebrafish. Understanding the function of these genes in fish will help clarify their roles in humans. The sequenced genome also permits “whole-genome/transcriptome” studies, in which, for example, the activity of all genes in response to an experimental treatment can be monitored. This can reveal previously unknown functions for genes, and lays the groundwork for understanding the gene networks that regulate processes from development to metabolism to fertility.
Several zebrafish-specific gene expression databases that provide information about when and where genes are active in developing embryos are available. Additional analysis tools have been developed to help parse and visualize expression data in useful ways.
Zebrafish mutants
In the 1990’s, two revolutionary large-scale random mutagenesis screens produced ~1,500 zebrafish mutants that are still used in labs across the globe (Nüsslein-Volhard et al., 2012). One example (which I will come back to) is the one-eyed pinhead mutant, which regulates a developmental pathway implicated in congenital heart defects and cancer. These mutants are used extensively to study development and disease. Additional mutagenesis projects have since been initiated and today thousands of mutations in genes affecting diverse biological processes—not just development—are available from strain distribution centers in the US, Europe, and China. In addition, the Zebrafish Model Organism Database ZFIN actively curates information on mutants and other resources.
Targeted gene deactivation tools for zebrafish
Random mutagenesis screens provided a major toehold into understanding how genes control biology (a fact celebrated by the Nobel prize committee). But as the field of genetics progresses and genome sequences become available, new hypotheses about specific genetic interactions arise and tools to deactivate specific genes are needed.
Targeted mutation has been demonstrated in zebrafish using zinc-finger nucleases, TALENs, and more recently CRISPR/Cas9. Another popular targeted gene deactivation method in zebrafish are short, modified oligonucleotides called morpholinos. When injected into zygotes, morpholinos complementary to a target mRNA inhibit its translation either by blocking initiation or splicing. (RNAi, in contrast, engages existing gene regulatory mechanisms to block activity and does not work easily in zebrafish.) Morpholinos are a convenient and fast way to deactivate genes without spending months generating mutants. Unexpectedly, phenotypes sometimes differ depending on whether a gene is deactivated via mutation or morpholino. This observation unlocked novel research into genetic compensation, a previously unknown mechanism of gene regulation (El-Brolosy et al., 2019).
Generating zebrafish transgenics
Creating transgenic organisms by introducing foreign DNA into the genome is useful for a variety of applications. For example, specific tissues such as blood cells or neurons can be fluorescently labeled, allowing researchers to observe changes to these tissues under different experimental conditions. Strategies for generating transgenic zebrafish are well established (Felker and Mosimann, 2016). One common and robust strategy uses a transposon called Tol2 to randomly integrate foreign DNA into the genome. Non-random integration into specific pre-determined regions is also possible using an alternative approach that exploits the bacteriophage integrase phiC31. The popular UAS/Gal4 and Cre/Lox gene regulatory systems have also been successfully implemented in zebrafish.
Developmental biology insights using lineage tracing in zebrafish
To highlight some of the features that make zebrafish useful, I’ll outline two recent lineage tracing approaches that led to important insights in developmental biology.
Microscopy-based lineage tracing
Over a period of 24 hours, the externally fertilized zebrafish zygote transforms into an organism with a clearly established body plan including head, eyes, and tail. Compared to the opaque mouse embryo, where similar processes occur over days inside the mother, the zebrafish is an ideal system to observe embryogenesis live. The origins, divisions, and movements of each of the thousands of cells in the developing zebrafish embryo have been tracked using microscopy and advanced computational approaches (Keller et al., 2008). This microscopy-based lineage tracing defined a high-resolution map of early embryogenesis in time and space, and was also used to precisely characterize developmental defects in the one-eyed pinhead mutant from the random mutagenesis screens. Microscopy-based lineage tracing can help illuminate how signaling pathways—such as those regulated by one-eyed pinhead—contribute to the “sculpting” of tissues and organs during embryo development.
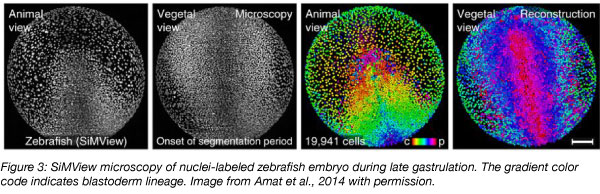 Single cell sequencing-based lineage tracing
Single cell sequencing-based lineage tracing
Recently, gene expression in all cells of the developing zebrafish embryo has been mapped using single cell RNA sequencing approaches (Harland, 2018). Several different methods have been developed, relying on established reagents such as CRISPR/Cas9, Tol2, and the high-quality sequenced zebrafish genome. One study profiled 38,731 cells from 694 embryos over multiple stages and reconstructed the patterns of gene activity and cell divisions that coordinate development of the body plan (Farrell et al., 2018). This study also showed that in the absence of the signaling pathway regulated by one-eyed pinhead, incorrect proportions of otherwise normal cell types are produced, rather than novel, abnormal cell types. Sequencing-based lineage tracing can help elucidate the gene networks that transform zygotes into adults and explain how signaling pathways control this process.
Additional zebrafish applications and conclusions
Zebrafish are also popular outside of developmental biology, and are particularly well-suited for neuroscience investigations. For example, neuronal activity can be tracked in whole brains of swimming, actively learning larvae, providing unprecedented views into cognitive processes like decision making (Kim et al, 2017). “Brainbow” fish lines have been developed to enable high-resolution mapping of the brain by uniquely labeling neurons. Zebrafish are also amenable to manipulation of brain activity: optogenetic approaches can be used to determine the consequences of activating or silencing specific neurons in living larvae. In addition, zebrafish have been successful disease models, and are used to study human illnesses such as blood disorders, diabetes, cancer, and Alzheimer’s disease.
Naturally, the best model organism depends on the biological question, and zebrafish are not ideal for every application. Disadvantages include their duplicated (i.e., redundant) genome, their long three-month generation time and labor-intensive maintenance compared to invertebrate model systems, and their resistance (so far) to facile CRISPR/Cas9-mediated insertion of foreign DNA.
Despite these caveats, several features of zebrafish—including the available genetic resources and transparency of the embryo—have proven useful in a variety of research applications. Lineage tracing studies highlight how these advantages can translate into insights in developmental biology, and advances in this field and others are likely to continue with help from zebrafish.
Many thanks to our guest blogger Katherine Rogers from the Friedrich Miescher Lab of the Max Planck Society.
 Katherine W. Rogers is a postdoctoral researcher in Patrick Müller’s group at the Friedrich
Katherine W. Rogers is a postdoctoral researcher in Patrick Müller’s group at the Friedrich
Miescher Laboratory of the Max Planck Society in Tübingen, Germany. She studies vertebrate
development using optogenetic approaches.
References
El-Brolosy, Mohamed A., et al. "Genetic compensation triggered by mutant mRNA degradation." Nature 568.7751 (2019): 193. PubMed PMID: 30944477.
Farrell, Jeffrey A., et al. "Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis." Science 360.6392 (2018): eaar3131. PubMed PMID: 29700225. PubMed Central PMCID: PMC6247916.
Felker, A., and C. Mosimann. "Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments." Methods in cell biology. Vol. 135. Academic Press, 2016. 219-244. PubMed PMID: 27443928.
Harland, Richard M. "A new view of embryo development and regeneration." Science 360.6392 (2018): 967-968. PubMed PMID: 29853675.
Keller, Philipp J., et al. "Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy." science 322.5904 (2008): 1065-1069. PubMed PMID: 18845710.
Kim, Dal Hyung, et al. "Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish." Nature methods 14.11 (2017): 1107. PubMed PMID: 28892088.
Nüsslein-Volhard, Christiane. "The zebrafish issue of Development." Development 139.22 (2012): 4099-4103.
Additional resources on the Addgene blog
- Read about zebrafish behavioral studies
- Download the fluorescent proteins 101 eBook
- Read our plasmids 101 blog series
Resources on Addgene.org
- Browse our zebrafish CRISPR plasmid collection
- Read our molecular biology reference
- Find plasmids for your research





Leave a Comment