This post was contributed by Neil J. McKenna and Bert W. O’Malley.
Survival of all organisms depends on efficient energy maintenance - through acquisition, storage, and utilization - and on self-propagation by reproduction. Both physiological processes are controlled by deliberate and compulsory actions instigated by the central nervous system signaling to peripheral effector organs, which then return information such as nutritional status. Within the animal kingdom, this information is relayed through factors (both of endocrine and dietary origin) that are diffused or actively transported from cells, traverse the body through the bloodstream, and eventually elicit their actions on other tissues. At the Nuclear Receptor Signaling Atlas (NURSA), one of our primary goals is to promote an understanding of how these processes are regulated at the cellular, tissue and organ level by nuclear receptors (NRs), their coregulators, and their physiological endocrine ligands.
Nuclear receptors and their ligands
NRs represent the largest family of transcription factors found in metazoans. The superfamily is comprised of 48 human and 49 murine members. NRs permit the integration and communication of such signals between central and peripheral organs because of their established roles as molecular sensors and governors of endocrine-hormone signaling. The success of members of this group stems in part from their ability to function as ligand-dependent sensors for a diverse set of fat-soluble hormones, vitamins, and dietary lipids. Included in this family are receptors for endocrine steroids (i.e., corticosteroids, progesterone, androgens, and estrogens), fat-soluble vitamins A and D, thyroid hormone, fatty acids, oxysterols, bile acids, and numerous xenobiotic lipids derived from the diet. Additional members of this family are called orphan receptors because their ligands remain unknown.
Together, NRs govern expression of genes involved in a broad range of reproductive, developmental, metabolic, and immune response programs. NRs also encompass one of the most successful targets for drugs currently available or being developed to treat a multitude of therapeutic indices, including hypertension, cancer, diabetes, cardiovascular disease, cholesterol gallstone disease, senescent illnesses, and the metabolic syndrome.
Coregulators
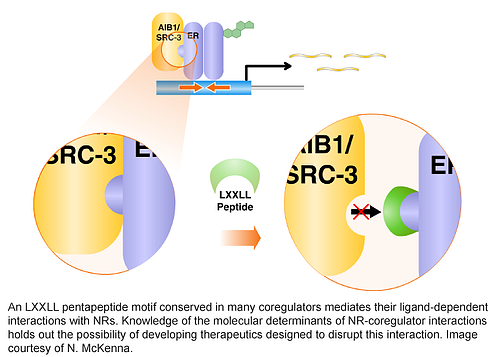
Coregulators constitute a group of some 400 molecules that have been shown to be required by one or more NRs for efficient and timely activation or repression of gene expression (see www.nursa.org for a curated list of coregulators). Coregulators can function as coactivators and corepressors depending upon a context defined by ligand, cell type, and promoter, organizing the expression of functional groups of genes involved in the execution of a specific regulatory regime (such as genes involved in metabolism, growth or cytokine action). Primary or core coregulators (those that interact directly with NRs) exist in steady-state complexes with secondary coregulator (or co-coregulator) partners. The coregulator ‘core’ complex is composed of a tightly-bound invariant group of proteins, whereas the more loosely bound co-coregulators associate with the core complex in a dynamic, regulated manner.
The fact that coregulators belong to distinct complexes may explain how more than 300 different coactivator proteins individually contribute to cell regulation in a coherent manner. Because coregulators exist as multiprotein complexes, a member of a single coactivator complex can serve as a rate-limiting conduit to control the actions of the whole complex. For example, the phosphorylation status of SRC-3 defines its association with other members of the complex, such as p300 and CBP histone acetyltransferases or CARM1 methyltransferase. This attendant signaling feature afforded by coactivator complexes suggests that coactivators may be integrators of multiple cell signaling systems. Activation of membrane receptors and signaling cascades conceivably allows the genome to sense the impact of the total environment on the cell.
The NURSA Consortium
Now in its third phase of funding, the NURSA Consortium represents a unique funding model in nuclear receptor research, and has generated a set of comprehensive datasets that collectively address many important and fundamental questions in nuclear receptor and coregulator research. Our Q-PCR datasets, for example, document the expression patterns of nuclear receptors across multiple different tissues in normal and disease states, their circadian regulation, as well as in important physiological states such as adipogenesis and macrophage activation. Our mass spectrometry-affinity purification proteomic profiling of coregulators in HeLa cells was the first comprehensive analysis of the interactome of these key regulatory molecules, and has provided clues to their previously uncharacterized cellular roles.
In the current phase of NURSA we are seeking to compliment these consortium-generated datasets with a systematic effort to compile NR- and coregulator-related global gene profiling datasets in to our data mining tool, Transcriptomine. We are currently nearing the completion of an 18-month effort to overhaul the NURSA website, the new version of which will feature a simplified user interface, expanded numbers of curated molecules, including 400 or so coregulators, and a greatly expanded set of links to datasets and reagents in the field. Additional information can be found in the Datasets and Reagents sections of the current NURSA website.
NURSA and Addgene
The great progress that has been made in the NR field over the last thirty years or so would not have been possible without the development of a vast number expression and reporter plasmids and the free exchange of these vectors between laboratories. The development of the transient transfection platform for assaying transcriptional activity, for example, was a key development in the functional characterization of NRs and coregulators, and although the reporter gene has over the years evolved from chloramphenicol acetyltransferase to luciferase, the basic assay remains widely used today.
The sheer abundance of these critical research tools is a double-edged sword, however, and has greatly increased the burden associated with identifying plasmids in the research literature and acquiring them in a timely manner. Addgene represents an excellent solution to the problem of sharing plasmids, providing researchers with a one-stop shop for plasmids and vectors, and making them available to all researchers for a reasonable fee. We at NURSA recognize the importance of this service to the NR research community and, accordingly, the new version of the NURSA website will feature direct links to listings of available plasmids for our curated NRs and coregulators. We look forward to growing this important relationship with Addgene for the benefit of the entire research community.
Thank you to our guest bloggers!

Neil McKenna, Ph.D., is Associate Professor of Molecular and Cellular Biology at Baylor College of Medicine and Editor-In-Chief of the Open Access MEDLINE-indexed journal Nuclear Receptor Signaling. Dr. McKenna edits the website of the Nuclear Receptor Signaling Atlas (NURSA) at www.nursa.org.

Bert O’Malley, M.D., is Tom Thompson Professor and Chair of the Department of Molecular and Cellular Biology, and Editor-In-Chief of the Open Access MEDLINE-indexed journal Nuclear Receptor Signaling.
Further Reading:
- Chawla, A., Repa, J. J., Evans, R. M.and Mangelsdorf, D. J. (2001). Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866-1870. (PubMed)
- Lonard DM and O’Malley BW. (2007). Nuclear receptor coregulators: Judges, juries and executioners of transcriptional regulation. Molecular Cell 27, 691-700. (PubMed)
- McKenna, N. J.and O'Malley, B. W. (2002). Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108, 465-474. (PubMed)
- McKenna NJ and O'Malley BW (2005) An interactive course in nuclear receptor signaling: concepts and models. Sci STKE. 2005, tr22.
Keep on reading! Check out Addgene's Plasmids 101 blog series.
Topics: Other, Miscellaneous







Leave a Comment