This post was contributed by Kevin Esvelt, a Wyss Technology Development Fellow at the Wyss Institute and Harvard Medical School.
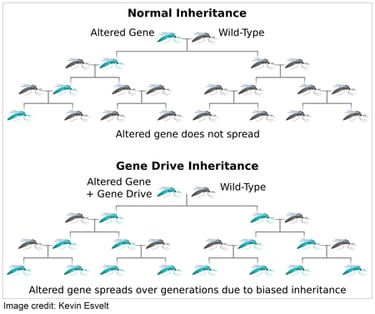 Scientists making transgenic organisms with Cas9 should be aware of the potential hazards of creating “gene drives” capable of spreading through wild populations. Whereas most genomic changes impose a fitness cost and are eliminated by natural selection, gene drives distort inheritance in their favor and consequently can spread even when costly.
Scientists making transgenic organisms with Cas9 should be aware of the potential hazards of creating “gene drives” capable of spreading through wild populations. Whereas most genomic changes impose a fitness cost and are eliminated by natural selection, gene drives distort inheritance in their favor and consequently can spread even when costly.
If even a single organism carrying a synthetic gene drive were to escape the laboratory, the drive could eventually spread through the entire wild population with unpredictable ecological effects. Because the consequences of such a mistake would necessarily extend far beyond the laboratory and seriously damage public trust in scientists, experiments involving potential gene drives should be conducted with extreme caution.
Why does making transgenic organisms with Cas9 risk creating a gene drive?
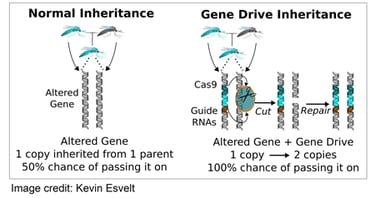 The simplest way to make a gene drive is to insert an endonuclease gene within its own cut site1. In heterozygotes, the endonuclease cuts the wild-type chromosome and its own gene is used as a repair template, thereby converting the heterozygote into a homozygote and ensuring that all future offspring inherit the endonuclease gene. As we pointed out last year2, delivering a DNA cassette encoding the cas9 gene and single guide RNAs (sgRNAs) with appropriate flanking homology into a germline cell can create an RNA-guided gene drive. In principle, this event could happen even if there isn't any homology available. Consequently, the best way to avoid accidentally creating a gene drive is also the easiest: don't use a DNA vector that encodes both Cas9 and sgRNA.
The simplest way to make a gene drive is to insert an endonuclease gene within its own cut site1. In heterozygotes, the endonuclease cuts the wild-type chromosome and its own gene is used as a repair template, thereby converting the heterozygote into a homozygote and ensuring that all future offspring inherit the endonuclease gene. As we pointed out last year2, delivering a DNA cassette encoding the cas9 gene and single guide RNAs (sgRNAs) with appropriate flanking homology into a germline cell can create an RNA-guided gene drive. In principle, this event could happen even if there isn't any homology available. Consequently, the best way to avoid accidentally creating a gene drive is also the easiest: don't use a DNA vector that encodes both Cas9 and sgRNA.
What if I want to work with gene drives?
Then do so! It's a tremendously promising field that could address many pressing global problems in health, agriculture, and conservation. But please be aware that there are many easy-to-implement confinement strategies that are robust to human error. Used in combination with standard barrier protocols, they can reduce the risk of accidental escape to a negligible level.
One major reason we chose to publish the concept and likely capabilities of RNA-guided gene drives in the summer of 2014 (after extensive discussions with biosecurity experts, ecologists, and national authorities) was to detail these robust and easy-to-implement confinement strategies2. Our hope was that anyone else who came up with the same idea or wanted to build a gene drive in a model organism would find our work and consequently use these precautions in their own experiments2,3. While that didn't work quite as well as anticipated4, greater awareness will certainly improve safety.
What safeguards and confinement strategies are available?
Molecular confinement involves building gene drives that can spread through populations of transgenic laboratory organisms but not wild organisms. For example, an sgRNA-only drive will spread exclusively through populations that already express Cas9 from an unlinked locus, while a Cas9+sgRNA drive targeting a synthetic sequence will only spread in transgenic laboratory populations with that sequence2. Both methods are easy to implement and have been tested in yeast5.
Ecological confinement involves performing experiments in a geographic area where escaped organisms won't be able to find mates2. For example, ongoing experiments attempting to build gene drives in tropical mosquito vectors of diseases such as malaria and dengue are currently being performed in regions that don't have resident populations of the relevant mosquito species.
Reproductive confinement involves working with laboratory organisms that can't reproduce with wild ones. For example, Drosophila lines with compound autosomes are completely infertile when mated to wild fruit flies6. It's also worth noting that gene drive experiments are less hazardous in organisms that seldom reproduce sexually because the drive must be much more efficient and minimally harmful in order to spread.
Barrier confinement seeks to keep the organisms in the laboratory. It varies by organism, but your local biosafety officer should be familiar with appropriate measures. Barriers should be a component of all gene drive confinement strategies, but they should not be relied on exclusively because historical studies of pathogen research have conclusively shown that barrier protocols are vulnerable to human error. And with gene drives, one mistake can be enough.
Reversal drives are designed to overwrite a previous gene drive and thereby undo the genetic changes driven by the earlier intervention2. While an initial reversal drive cannot restore the exact original sequence, it can restore the original protein-coding sequence using a recoding strategy; a subsequent drive can restore the wild-type sequence (save for the residual sgRNAs and possibly cas9 gene). An immunizing reversal drive is a variant that also spreads through the wild population and immunizes it against the first drive. Laboratories interested in building candidate gene drives intended for eventual release should consider building an appropriate immunizing reversal drive at the same time to mitigate the potential effects of an accidental release3.
Which gene drive confinement strategies should be used?
There are ongoing efforts to develop formal guidelines to answer this question. Until then, using multiple confinement strategies is strongly advised. The effects are multiplicative, and there are very few gene drive experiments that can't be performed with two or more confinement methods. Since molecular, ecological, and reproductive confinement typically require very little effort to implement (depending on the species in question), why not use them whenever applicable?
Against this, consider the cost of an accidental release. Science relies on popular support, which in turn depends on widespread trust in scientists to develop new technologies with care and humility. Nothing would damage that trust more than an accidental gene drive release, which would represent proof positive that at least some scientists could not be trusted to handle such a powerful technology responsibly. An accidental release would be particularly dangerous for molecular biology and genetics because the underlying advance enabling RNA-guided gene drives is Cas9 genome editing.
Finally, gene drives are an inherently collective technology: because they necessarily alter the shared environment, they must never be used without popular support2. An accidental release could delay real-world applications against scourges such as malaria and dengue for many years. Since more than a thousand children died of malaria today, let's not risk a potential way to dramatically reduce that number for the sake of laboratory convenience.
Disclaimer: Recommendations concerning the appropriate degree of confinement are those of the author and do not represent a formal stance taken by Addgene or its staff. Additional information about gene drives can be found in the referenced publications.
Thank you to our guest blogger!
 An inventor of technologies that harness evolution, Kevin Esvelt studies ways of using molecular tools to alter populations of organisms and associated ecosystems. Because the environment is shared by everyone, he is deeply concerned with ensuring that gene drives and other collective technologies are developed safely and transparently. For more information, visit www.sculptingevolution.org.
An inventor of technologies that harness evolution, Kevin Esvelt studies ways of using molecular tools to alter populations of organisms and associated ecosystems. Because the environment is shared by everyone, he is deeply concerned with ensuring that gene drives and other collective technologies are developed safely and transparently. For more information, visit www.sculptingevolution.org.
References:
- Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). PubMed.
- Esvelt, K. M., Smidler, A. L., Catteruccia, F. & Church, G. M. Concerning RNA-guided gene drives for the alteration of wild populations. eLife e03401 (2014). doi:10.7554/eLife.03401. PubMed.
- Oye, K. A. et al. Regulating gene drives. Science 345, 626–628 (2014). PubMed.
- Gantz, V. M. & Bier, E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science aaa5945 (2015). doi:10.1126/science.aaa5945. PubMed.
- DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M. & Church, G. M. RNA-guided gene drives can efficiently and reversibly bias inheritance in wild yeast. bioRxiv 013896 (2015). doi:10.1101/013896. bioRxiv.
- Fitz-Earle, M., Holm, D. G. & Suzuki, D. T. Genetic Control of Insect Populations: I. Cage Studies of Chromosome Replacement by Compound Autosomes in Drosophila melanogaster. Genetics 74, 461–475 (1973). PubMed.
More CRISPR-Cas9 Resources:
- CRISPR 101: a blog series on the basics of CRISPR-Cas technology
Topics: CRISPR, CRISPR Biosafety






Leave a Comment