This post was contributed by guest blogger Cameron MacPherson at the Institut Pasteur
CRISPR software and the piñata effect
Two years ago I was a part of a group (Biology of Host-parasite Interactions, Institut Pasteur, Paris) that changed genome editing in the malaria community for the better (Nat. Biotechnol., 2014). Given the timing, it shouldn’t be a surprise that the CRISPR system was involved. Today, that same laboratory enjoys a successful edit rate of over 90% in their work editing the genome of Plasmodium falciparum (the parasite that causes malaria). I attribute their success to technical expertise, thoughtful single guide RNA (sgRNA) design, and the abnormally low GC content of the Plasmodium falciparum genome. To put this last point into perspective, the Plasmodium falciparum genome contains only 0.66 million targetable NGG PAM sites whereas the human genome has about 300 million. With such a sparsely targetable genome, off-targeting is less of a worry and on-targeting likely more efficient.
These insights are hard to appreciate without computational support. Indeed, rational sgRNA design is not possible without relying on some kind of pre-analysis. At the end of 2014, I began developing software to make sgRNA design accessible to all. At the time I thought there was room for improvement and a year later it became quite clear that others thought the same. Since January 2013 there have been 33 CRISPR software tools published and documented by OMICtools. It must be confusing for a newcomer to decide on the right software to use. With so much choice the question I have is, how did we get here in the first place?
How CRISPR innovation affected software development

CRISPR, like the miRNA bubble before it, offers an interesting view into how rapid innovation affects the research community. I’ve likened it to the celebration surrounding the piñata. From the perspective of those building the piñata, their role is to fill the prop with something of value. They are reviewers of content. On the other hand, the goals of the piñata beating party goers are altogether different. They are filled with blind anticipation. Their trust in the quality of the content is implicitly defined by their trust in the piñata builders - the reviewers. After some physical exertion on the part of the party goers, the piñata bursts and its contents spill to the ground. There is something akin to a frenzy as the crowd tries to weigh and determine the value of every sweet or toy strewn across the floor. In the final wake it all settles down and the value of each item changes from what the reviewers so painstakingly assessed to something a bit more representative of the communities’ opinion. The trouble is, with so much choice, in those few moments after the piñata burst, value was subjectively ascribed relative to the basic needs of each individual; the community as a whole has yet to settle on any true kind of value. I’ve dubbed this the “Piñata Effect”, it is the disconnect arising between those reviewing the contents of the piñata and the audience blindly receiving them; it results in highly similar content and the absence of iterative design; it is caused by not having the time to assess public response; and, it is the stage I think we are currently at in the CRISPR software space. The CRISPR research community hasn't had a chance to develop a consensus on the best CRISPR software tools yet.
As a genome editing tool, the CRISPR/Cas9 technology has been surrounded by a whirlwind of research activity and development since its inaugural year in 2012. That’s just 3 to 4 years ago. It is a bubble of innovation (a piñata), and software development has caught up with it. The peer review process has been challenged with many closely spaced CRISPR software submissions. With few prior publications to go on, a submitted manuscript could only have been viewed as a significant improvement. As a result, the software piñata has been filled with slight variations on a theme without many resources to review them. Collectively, the pool of CRISPR software embodies solutions that facilitate most experimental applications in CRISPR engineering. When compared to that collective utility, it’s easy to fault the lack of features in a single application. The question is then, should we use all tools as some kind of a meta/Frankenstein app? Or perhaps there is a clear winner, is there something that we can invest our time into learning and extract the most value? This is only one of many decisions faced by those engaging in CRISPR design, but it is a significant hurdle made worse by an ever increasing marketplace. Since 2013 about 11 tools have been added every year.
Breaking down the barriers: My view on current sgRNA design tools
The goal of this post is to provide some insight into available CRISPR software tools, what problems various tools are trying to solve, and finally how we might proceed in the future. When we think of designing experiments using CRISPR, there are two major areas where software can help. The foremost handles the design of sgRNA and represents the lion’s share of the currently available tools (we will focus on these tools in this blog post). The second major area deals in post-experiment quality control, a good example is CRISPR-GA. These tools assess repair events by type (NHEJ/HDR) and track indels at a single locus, usually the target site. Multi-locus asseEENdb database
ssment would be required to fully evaluate off-targeting, but currently no software has been designed for this purpose. This second area is extremely important and given the lack of attention to it, it is an obvious place to focus further development toward. Quality control is, however, not a focus of this blog post.
I separate sgRNA design tools into database and de novo solutions. Database tools allow us to view and get a sense of which sgRNA designs have previously worked and under what conditions. Such resources could be highly valuable for automated rational design. The first of only three such databases, EENdb (published 1 Jan 2013, no longer available), is a simple catalogue of reported sgRNA designs. The second database, CrisprGE (published 27 Jun 2015), is noteworthy for its broad scope, curated content, and ease of access. The third, WGE (published 15 Sep 2015), is more similar to EENdb than CrisprGE, but is also a good example of how database tools can be used to aid design. These databases are still young and will require a collaborative effort from the community in order to succeed.
With respect to de novo sgRNA design, I again separate it into two categories. On the one hand we have software tools that apply some approximate or sensible rules to determine the value of one sgRNA design over another. On the other hand some software tools make use of empirically derived and/or prior knowledge to inform on the qualities of what determines good sgRNA design. Incidentally, the first software for sgRNA design (by Hsu et al., 2013) used a combination of both empirical and approximate methods. The details of their scoring function can be found in their online tool’s documentation. Since Hsu et al., 2013, other tools have incorporated new parameters such as the Doench-Root score (by Doench et al., 2014) into their scoring functions. Many tools have also opted to mix-and-match different scoring algorithms and parameters. This tapestry of ideas is what makes choosing the right tool confusing and it means that there is no one-size-fits-all approach. You will have to select a tool based on your project and not general opinion.
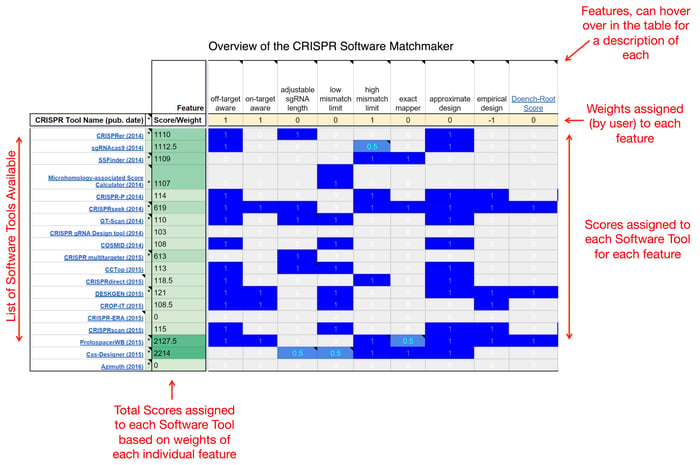
I could spend a lot of time running through each tool in detail, but that is already covered in the published articles. Instead, I have broken down the tools into the individual features that make each tool unique. The CRISPR Software Matchmaker is composed of these features and enables you to select the tool(s) based on your project needs. There are 8 major categories found in the table, describing everything from basic to advanced functionality as well as how the user is expected to interact with the tool (a screen shot and overview of the table can be found above). These categories are further discussed below (definitions for all terms are also available in the table):
- Basic functions: These functions embody the main goals of the software and should be the first place you look to determine a tool’s suitability. Functions of this category are the most common between tools. Examples: “single-target design”, “multi-target design”, “off-target aware”, “high mismatch limit”, “approximate design”, “empirical design”, “single-PAM design”, and “multi-PAM design”. Trends: By the end of 2014, tools began moving away from designing exclusively for NGG PAM targets and began allowing for arbitrary PAM definitions. These newer tools may still prove somewhat useful for the latest NTT targeting Cas protein, Cpf1. Separately, on-target efficacy is still a concept that only a few tools are trying to solve.
- Advanced functions: These functions are not entirely necessary but should be considered extremely useful, depending on design goals. Examples: “feature aware”, “SNP aware”, “secondary structure aware”, and “microhomology aware”. Trends: Fewer tools are being released with advanced functionality, it seems these kinds of features are delegated to other more suitable software such as ApE or commercial workbenches such as MacVector or Benchling.
- Utility functions: These functions help speed up the sgRNA design process by removing repetitive tasks and/or by providing features to help with the post-design process such as primer and plasmid design. Examples: “multiplex design”, “multi-method design”, and “single-method design”. Trends: The most common utility functions are batch design or multiplex features. However, tools aiding in primer design are more and more common.
- User interaction: Software design elements that fall into this category describe how the user is expected to interact with the software. This category is important for users wishing to select tools based on their comfort level with operating a computer. Examples: “offline”, “online”, “CLI”, and “GUI”. Trends: Online tools dominate the software space but generally rely on less powerful algorithms to detect off-targets. The few offline tools available mostly work on any computer but generally require working knowledge of the command line or scripting.
- Input flexibility: Software tools require some kind of data input in order to generate results. While different types of input are more user-friendly than others, this is of less concern than data output as different input types can be readily converted. Examples: “organism”, “sequence”, “identifier”, and “load”. Trends: Sequence based input is by far the most common with most tools also requiring that the user specify an organism.
- Output diversity: Different tools provide results to the user in different formats. This can have a large impact on downstream results. Examples: “HTML”, “visual track”, “plot”, “tabular”, “interactive”, and “save”. Trends: Most tools provide tabular HTML formatted output. Surprisingly only 2 tools provide FASTA output. A few notable tools allow the user to save the results and return later or share them.
- Community exclusivity: Different communities develop software for their own needs. This often results in software design that is applicable to only one, or a subset of organisms. In only a few cases the software is organism agnostic. Most tools are exclusive to one or a subset of organisms. Examples: “organism agnostic”, “model organisms”, “small genome”, “large genome”, and “organism biased scoring”. Trends: Many tools can handle genomes of any size and are only limited by the time it takes authors to add a genome to the tool’s repertoire. Organism agnostic tools are the better solution but are often packaged as offline tools requiring expertise in one programming language or another.
- Supported organisms: This section lists the genomes supported by each tool. The genomes are denoted by organism name and genome assembly version. The names in the table appear as they do in the software. Most sgRNA design tools require the user to specify an organism. They do this because they rely on pre built indices or databases in order to find and present the results to you as soon as possible. Some tools don’t require the organism to be specified and are truly organism agnostic. Other tools require an organism to be specified but also allow you to build your own database; these tools will be marked as organism agnostic and have currently available organisms listed in this section.
CRISPR software advice for specific users
You will find that some tools cater to specific use-cases through the way their algorithms were developed or by their focus on specific organisms. For this reason I generally recommend that laboratories that have already attempted several CRISPR experiments evaluate the predictive power of each tool they are interested in by comparing their past results to the designs suggested by each tool. For those who are entering sgRNA design for the first time, it is best to choose a tool based on immediate needs and re-evaluate after several experiments. Alternatively, the databases mentioned above or, even better, first-hand knowledge from laboratories employing CRISPR on the same organism could be used as a proxy for this last approach.
Top tool for first timers: You need to learn the language and what the parameters represent. The best learn-by-doing tool is E-CRISP. The authors have made a huge effort so that their tool is both didactic and functional. It is also the only tool I have come across to offer classes of parameters based on the type of CRISPR experiment being pursued. You should also look at the glossary section of the CRISPR Software Matchmaker, it provides a list of terms I think are important to define for the CRISPR software space. It is unlikely that you will find these terms anywhere else, because I developed them for this post, but they should give you an idea of what to look out for.
Top tools for bioinformaticians: For flexibility in your own analysis, you need access to raw data, the target sites, off-target sites, scores and the statistics that go into calculating them. For this, some online tools such as Cas-OFFinder will suffice. Cas-OFFinder provides a bare-bones data dump of ALL strings in a genome using any IUPAC encoded pattern at any edit distance. It won’t, however, compute any score other than the edit distance. Added functionality is left to you to implement. You can also download the software for use on any machine with OpenCL enabled hardware. This OpenCL dependency is a severe limitation, but the tool is fast enough that, if you’re engaged in heavy CRISPR design (I’m talking screens and/or very large genomes), then it is worthwhile buying a dedicated machine. On the offline front, there is a Python script called SSFinder, but I find its implementation to be too slow for practical use. The R based package CRISPRseek offers great utility and coupled with Bioconductor should be the first choice for anyone already familiar with R. For the Java enthusiasts, take a look at sgRNAcas9.
Top tools for transferring CRISPR technology to a new organism: For obvious reasons you can rule out any tool that limits you to a subset of specific genomes. These are most of the tools, but the good news is that you only need one. ProtospacerWB was developed with the purpose of applying CRISPR technologies to new organisms and will help you whether you have a full assembly or not. It is an offline tool, but comes with a graphical user interface.
Best strategy for labs: Define what you need the software tool to do before going shopping for one. Use the CRISPR Software Matchmaker to select the best tool based on your needs. Refine your criteria. Repeat until you have found the best tool. Do several experiments and use the results to re-evaluate all tools. Report your findings to help everyone else.
CRISPR technology has reached into so many different communities that it is all but impossible to fairly judge individual tools. I believe at this time it is only possible to objectively break the tools into their individual offerings and allow you to select them based on your needs. We will likely begin to see a contraction of available tools with more and more features being integrated into so-called genome editing workbenches such as Benchling (commercial, online), DESKGEN (commercial, online), or ProtospacerWB (academic, offline). While it remains to be seen, I believe the future of CRISPR software is promising. Rapid innovation in this space has put us in a good position to step back and cherry pick the features that will actually make a difference in our experiments and lives. To do this effectively, there needs to be good communication between developers and end users.
Thanks to our guest blogger Cameron MacPherson!

Cameron R. MacPherson is Lead Data Scientist for the Milieu Intérieur project, Institut Pasteur, Paris. You can follow him on Twitter @CMacPhD.
References
1. Ghorbal, Mehdi, et al. "Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system." Nature biotechnology(2014). PubMed PMID: 24880488.
2. Hsu, Patrick D., et al. "DNA targeting specificity of RNA-guided Cas9 nucleases." Nature biotechnology 31.9 (2013): 827-832. PubMed PMID: 23873081. PubMed Central PMCID: PMC3969858.
3. Doench, John G., et al. "Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation." Nature biotechnology (2014). PubMed PMID: 25184501. PubMed Central PMCID: PMC4262738.
Resources at Addgene
Get Tips on How to Design a gRNA
Read Our CRISPR Guide to Catch up on All the CRISPR Basics
Read Other Blog Posts about CRISPR
Find validated gRNAs
Topics: CRISPR, Other CRISPR Tools







Leave a Comment