As a plasmid repository, we are no strangers to cloning. If you’re reading this, then chances are you have been in the cloning trenches yourself. Modular cloning, or MoClo, is a systematic cloning strategy designed by synthetic biologists to perform large scale, one-tube cloning reactions. In this blog, we’ll cover the basics of what MoClo is and what types of experiments it enables.
What is modular cloning?
Modular cloning, or MoClo, is an organization scheme for multi-part DNA assembly. Simply put, it assembles individual DNA components into larger units or circuits with functional relevance following a set hierarchy.
MoClo originally developed from Golden Gate Cloning, in which a type IIS restriction enzyme and a ligase are used to assemble multiple DNA fragments together in a one-tube reaction. Even though most MoClo reactions use Golden Gate cloning, not all Golden Gate cloning is MoClo. That’s because MoClo specifically refers to the organizational schema described above, and not any particular cloning technique. Though rarer, you can find MoClo that relies on Gibson and other non-Golden Gate cloning methods.
MoClo’s organization schema
MoClo can be broken down into three hierarchical levels: individual DNA parts, transcriptional units, and multigene constructs. Let’s start by examining individual parts and transcriptional units.
Individual parts are defined DNA sequences with specific functions. Most individual parts have characterized activities with predictable behaviors. While a gene is the most obvious example of an individual part, this category also includes promoters, UTRs, terminators, etc. Once selected, individual units can be combined into…
Transcription units (TUs), which are functional units composed of multiple parts transcribed together. They can express one or more gene(s), and typically begin with a promoter and end with a terminator. A simple TU could be comprised of just three parts: promoter, gene, and terminator.
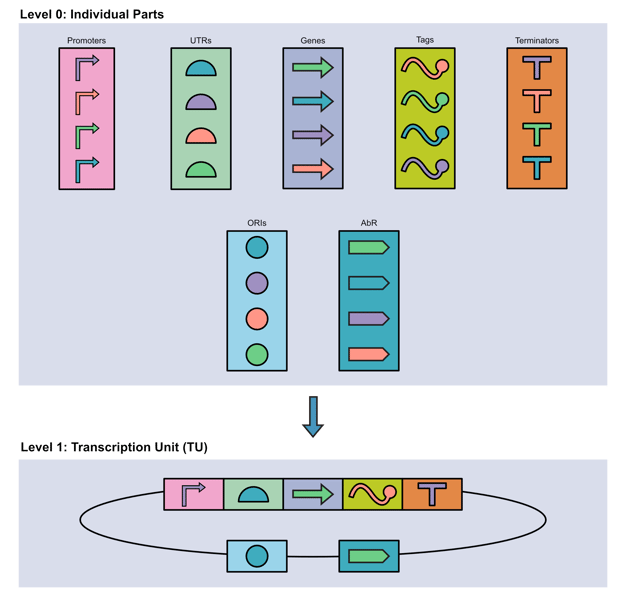
|
| Fig 1. MoClo enables individual parts to be assembled into larger functional units which are transcribed together. Image courtesy of Robert Hurt. |
Understanding how individual parts can be assembled into TUs is key to understanding MoClo. By a similar principle, TUs can be further complexed into multi-TU constructs, known as multigene constructs.
Construction of multigene constructs, through multi-TU assembly,happens via three steps:
- Level 0: Cloning of basic parts (promoters, UTRs, genes, tags, etc.) into MoClo-compatible vectors.
- Level 1: Assembly of multiple Level 0 parts into TUs.
- Level 2: Assembly of multiple Level 1 TUs into multigene constructs.
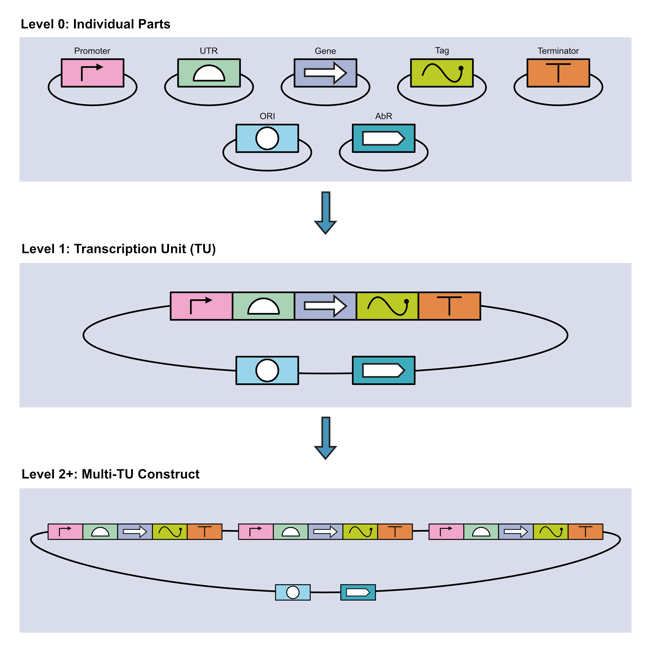 |
| Figure 2: The three assembly levels of MoClo. Image courtesy of Rob Hurt. |
What makes MoClo systems so powerful is their modularity — many parts can be assembled into many combinations. Even with current technologies, it can be extremely hard to predict how a single TU, or even an individual part, will behave in a cell. For unpredictable cases, MoClo provides two answers: first, sets of parts with characterized behaviors are available from Addgene and other sources; second, characterizing a part or optimizing a TU is much easier when you can quickly test that part in different contexts and plug different characterized parts into a TU!
What is modular cloning useful for?
All of this sounds really neat — but when is this type of large-scale assembly useful, you may ask? You might be surprised by how multi-purpose it can be. Let’s review a list of MoClo applications.
Genetic circuits
Most things in biology are not so simple, and engineering complex cellular behaviors requires making complex manipulations to cells. For example: in a cell there are proteins which interact with other proteins and with DNA, and these complexes regulate the transcription of specific genes. What if one of those genes coded for a protein that regulated its own transcription, or the transcription of another gene? Now you have a genetic circuit! Genetic circuits are multigene systems whose protein products can interact with the DNA that encodes the protein itself or other genes within the circuit. These interactions yield a cellular phenotype, translational output, etc., which can support a larger cellular engineering goal, several of which are described below. Multigene circuits assembled through MoClo can allow you to study natural genetic circuits in the cell.
Natural and synthetic product production
A major goal of synthetic biology is to efficiently produce biofuels, drug precursors, etc., microbially or in cell-free systems. These pathways typically involve multiple steps in which a precursor molecule is made, modified (sometimes many times), and isolated afterwards. To engineer these production pathways, MoClo has been used to assemble multigene constructs capable of performing production and subsequent necessary modification of these chemical products. The flexible nature of MoClo allows users to test a variety of different orientations and components within the system to maximize production potential. For example, the best promoter for a given peptide can easily be tested by generating Level 1 TUs with unique promoters, and the ideal phosphatase (or any modifying enzyme) to pair with it can be queried by testing any number in the Level 2 assembly. These pathways can require a lot of optimization - which is why MoClo is so attractive! It allows researchers to assemble and test many variables all at once.
CRISPR
CRISPR arrays and libraries depend on many gRNAs delivered with a relevant Cas nuclease. MoClo can be used to assemble multiplexed gRNA systems as well as additional components such as selectable markers, promoters, enhancers, etc.
MoClo is especially useful for more complex CRISPR experiments (e.g., many guides/parts delivered) in certain model organisms. In mammals, most CRISPR libraries are assembled and delivered virally, but these reagents and strategies are not available to those working with yeast or plants. Using MoClo, or better yet, a pre-made MoClo kit, is a prime choice for assembling an array of targeting constructs and delivering them to these organisms via another optimized platform.
Gene Stacking
As the name implies, gene stacking is the use of a combination of multiple genes to achieve a desired effect. This is commonly used in plant biology to achieve traits such as disease resistance. MoClo is a great tool to deliver many combinations of different candidate genes at once to identify the best cocktail of genes when the relationship between genotype and phenotype is best described with “it’s complicated.”
Applying MoClo
Interested in using MoClo? You don’t have to start from scratch! As you can imagine, there is an array of distinct goals within metabolic engineering, synthetic product production, and gene editing that use MoClo, for which many different MoClo kits have already been established. Keep an eye out for our next blog on choosing a MoClo strategy and kit if you want to learn more about this cloning technique!
Many thanks to Rob Hurt for his help on this piece. Rob is postdoc in Peter Schultz’s group at Scripps Research, where he works on genetic code expansion.
References and Resources
References
Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011 Feb 18;6(2):e16765. doi: 10.1371/journal.pone.0016765.
Hahn, F., Korolev, A., Sanjurjo Loures, L. et al. A modular cloning toolkit for genome editing in plants. BMC Plant Biol 20, 179 (2020). https://doi.org/10.1186/s12870-020-02388-2
Resources on Addgene.org
Resources on the Addgene blog
Topics: Plasmids 101, Plasmids





Leave a Comment