If you were born before 1985, you might remember going to the store and buying CDs when you wanted to hear a piece of music. It was tough to share or remix the songs, the CDs could get scratched over time, and it was difficult to keep track of growing collections of music. I still have a shelf full of CDs that have been gathering dust since the digital music revolution hit. I predict a similar revolution for antibodies. Recombinant antibody technology makes it easy for scientists to share, remix, store, and track these essential components of the scientific toolbox.
What are Recombinant Antibodies?
Antibodies are used by scientists around the world to detect, purify, quantify, deplete, and visualize proteins of interest (Greenfield, 2014). Traditionally, they have been made as either polyclonal antibodies or monoclonal hybridomas, but those techniques have several drawbacks. Polyclonal antibodies produced in animals may be variable among animals and bleed dates and even clonal hybridoma cell lines can produce more than one mAb (Bradbury et al., 2018). Hybridomas can lose gene expression of the mAb-encoding genes or fail to revive after cryopreservation (Bradbury & Plückthun, 2015a, b). Therefore, researchers from over 100 scientific institutions have proposed a shift to recombinant DNA-based antibody technologies (Bradbury & Plückthun, 2015a).
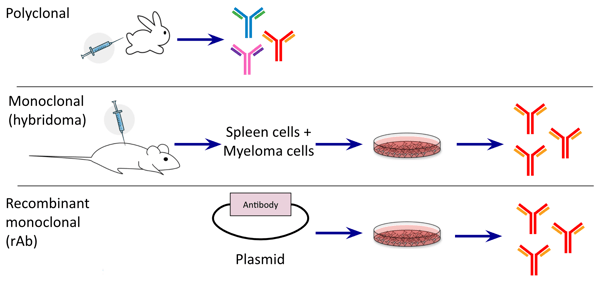 |
| Figure 1: (Top) Polyclonal antibodies are produced in animals and consist of a mixture of antibodies recognizing many different epitopes. (Middle) Monoclonal antibodies produced from hybridomas are usually a single antibody recognizing one epitope. (Bottom) Recombinant monoclonal antibodies are encoded in plasmids and produce a single antibody recognizing one epitope. |
Recombinant antibodies are monoclonal antibodies that are generated in vitro using synthetic genes that are typically expressed from a plasmid or from an integrated sequence in a stable cell line. The genes encode the heavy chain and light chain for the antibody and when translated into protein will assemble into a fully functional antibody. These antibodies can be used just as you would use antibodies made from animals or hybridomas.
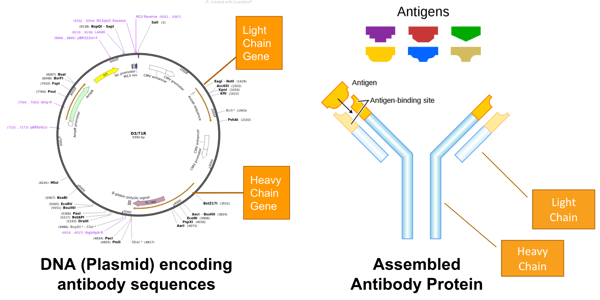 |
| Figure 2: (Left) Figure 2: (Left) The light chain and heavy chain genes of the antibody are encoded in a plasmid. (Right) An assembled antibody protein binds an antigen. Image from Fvasconcellos. |
How are Recombinant Antibodies Made?
To make recombinant antibodies, you first need to know the sequence. Scientists are getting better and better at identifying and cloning the genes for antibody expression. If starting from a protein sample (for instance, affinity purified antibodies from a patient’s blood), scientists can use mass spectrometry to determine the amino acid sequences of the antibodies and then synthesize the genes that encode those amino acids (Tran et al., 2016). Each antibody would then be tested for antigen binding. If starting from an established hybridoma line, the antibody can be converted into recombinant form through DNA sequencing of the line and subsequent cloning of the antibody chains (Crosnier et al., 2010; Andrews et al., 2019). Finally, scientists can perform the entire process of antibody selection and maturation in vitro by selecting for antigen binding from libraries of recombinant antibodies. The selection process is often performed via phage display, yeast display, ribosome display, or mammalian display (Tsuruta et al., 2017). In vitro selection allows scientists to create antibodies to targets that may not work well in animals because of similarities to host proteins.
After the antibody of interest has been cloned into an expression plasmid, the plasmid can be introduced into host cells, such as bacterial, yeast, or mammalian cells, for antibody production and subsequent purification.
Academic laboratories and companies have already begun creating plasmids encoding affinity reagents. These include conventional heavy and light chain recombinant monoclonal antibodies as well as other forms of antibody-based (e.g., nanobodies, ScFVs) and non-antibody based affinity reagents (e.g., monobodies, DARPins) (Helma et al., 2015).
The Many Benefits of Recombinant Antibodies
Recombinant antibodies offer numerous advantages over polyclonal antibodies and traditional hybridomas.
First, the long-term stability, consistency between batches, and molecular definition of recombinant antibodies are essential for good reproducibility. Non-recombinant antibodies are notorious for being the source of irreproducible data (Begley & Ellis, 2012; Baker, 2015). This contributes to an enormous amount of wasted money and time, with an estimated $350M/year lost on low quality antibodies in the US alone (Bradbury & Plückthun, 2015a). With molecularly defined antibodies that do not change over time, scientists will know exactly what antibody they are using and any validation experiments performed will be useful in perpetuity.
Second, plasmids are easy to store and share, making recombinant antibodies the most practical choice. Hybridomas are much tougher to ship, often lack sequence data, and may genetically drift over time. Antibodies made from animals are limited in supply and producing antibodies from plasmids is much kinder to our animal friends!
Finally, if we want better antibodies, we need to empower the scientific community with the tools to engineer them. Given the ease with which scientists can now improve genetically-encoded research tools, it is shocking that the plasmids and sequences for antibodies are not shared. For instance, through open plasmid sharing, the CRISPR community has created hundreds of useful variants of Cas9. By providing access to the plasmids encoding antibodies, scientists will be able to make higher affinity antibodies, modify antibody function, improve antibody stability, and design tools that we haven’t even imagined yet.
References and Resources
References
Andrews NP, Boeckman JX, Manning CF, Nguyen JT, Bechtold H, Dumitras C, Gong B, Nguyen K, van der List D, Murray KD, Engebrecht J, Trimmer JS (2019) A toolbox of IgG subclass-switched recombinant monoclonal antibodies for enhanced multiplex immunolabeling of brain. eLife 8: . https://doi.org/10.7554/elife.43322
Baker M (2015) Reproducibility crisis: Blame it on the antibodies. Nature 521:274–276 . https://doi.org/10.1038/521274a
Begley CG, Ellis LM (2012) Raise standards for preclinical cancer research. Nature 483:531–533 . https://doi.org/10.1038/483531a
Bradbury A, Plückthun A (2015) Reproducibility: Standardize antibodies used in research. Nature 518:27–29 . https://doi.org/10.1038/518027a
Bradbury AM, Plückthun A (2015) Antibodies: validate recombinants once. Nature 520:295–295 . https://doi.org/10.1038/520295b
Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, Bladen CL, Jones B, Aldred SF, Bestagno M, Burrone O, Maynard J, Ferrara F, Trimmer JS, Görnemann J, Glanville J, Wolf P, Frenzel A, Wong J, Koh XY, Eng H-Y, Lane D, Lefranc M-P, Clark M, Dübel S (2018) When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. mAbs 10:539–546 . https://doi.org/10.1080/19420862.2018.1445456
Crosnier C, Staudt N, Wright GJ (2010) A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biology 8:76 . https://doi.org/10.1186/1741-7007-8-76
Greenfield, E.A. (Eds.), Antibodies: a Laboratory Manual, 2nd Edition. 2104. Cold Spring Harbor Laboratory Publications, New York.
Helma J, Cardoso MC, Muyldermans S, Leonhardt H (2015) Nanobodies and recombinant binders in cell biology. Journal of Cell Biology 209:633–644 . https://doi.org/10.1083/jcb.201409074
Tran NH, Rahman MZ, He L, Xin L, Shan B, Li M (2016) Complete De Novo Assembly of Monoclonal Antibody Sequences. Sci Rep 6: . https://doi.org/10.1038/srep31730
Tsuruta LR, dos ML, Moro AM (2018) Display Technologies for the Selection of Monoclonal Antibodies for Clinical Use. In: Antibody Engineering. InTech
Additional resources on the Addgene blog
- Read our Antibodies 101: Introduction to Antibodies blog post
- Learn more about hybridomas
- Watch our Antibodies 101: What is an Antibody? animation
Resources on Addgene.org
- Check out Addgene's antibody plasmid collection
Topics: Antibodies







Leave a Comment