It is a well known fact if a multicopy variant of a gene exists, there must be assays that researchers can use to quantify said copies. They do so using copy number variation (CNV) assays. There are several varieties of these, but in this blog post, we're going to focus on PCR-based applications (qPCR and ddPCR.)
qPCR
Quantitative/real-time PCR (qPCR) has been used for copy number assays for years. qPCR works like regular PCR, just with signaling molecules added to the mastermix that are read after each cycle. The raw data is then normalized to a reference sequence present in the same amount in all samples (usually to a ubiquitously expressed, one-copy ‘housekeeping’ gene), and quantified relative to a 'normal' sample with known copy number set as the expression baseline. Copy number is then determined by comparing results to a standard curve. For CNV assays, it is recommended to use a reference sample with two copies of the gene (Ma, 2014.) As with all qPCR assays, samples, standard curves, and normal controls must be run in triplicate.
The nice thing about qPCR is that since it measures relative quantification - that is, since you’re looking at the amount of DNA in your sample relative to your normal control - you can set up your calculations so that your assay output number (RQ value) matches the copy number of your sample. In that case, an RQ value of one (or, more realistically, between 0.80-1.20) would mean you have one copy of the gene; an RQ value of 2 means you have two copies, and so on and so forth. This also serves as a light technical control - an RQ value of 1.5 means you have either made a pipetting error or an incredible discovery. The range of RQ values used to call copy numbers should be as small as reasonably possible and set using the variance seen within multiple runs of the standard curve.
Signaling molecules: Sybr Green and Taqman
qPCR most often uses either Sybr Green or Taqman for signaling. Sybr Green is a fluorescent dye that works by binding to all double-stranded DNA, while Taqman uses an intercalating dye, with a fluorescent probe and quencher to quantify specific amplicons.
Sybr has high background noise, due to the large amount of dsDNA in most starting materials, and is not particularly sensitive. The fluorescent signal can be affected by AT-rich DNA and the length of the amplicon and it has a higher rate of false positives (Colborn, 2008; Harridan, 2015). Sybr, therefore, cannot be used to accurately call copy numbers, though it has been shown to identify trends correctly (Muhamad, 2011; Providenti, 2006). Its biggest advantage over Taqman is price point, which should be carefully balanced against the limitations of the assay.
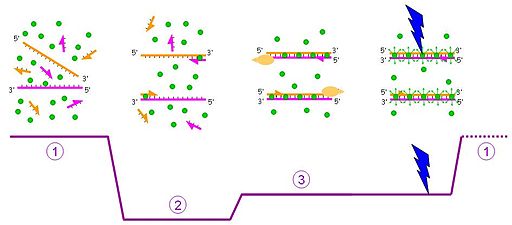
|
| Fig. 1: A schematic representation of Sybr Green chemistry. Blue arrows represent fluorescent signaling in the presence of dsDNA. Image courtesy of: Ygonaar |
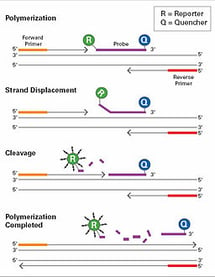
Taqman is more sensitive than Sybr, with less background noise, and is not affected by AT content or length of the amplicon. As the probes can have several different markers, it can also be used to run multiplex assays, allowing the reference sequence and the gene of interest to be amplified in the same reaction. Taqman’s increased sensitivity allows for a more confident determination of copy number, albeit within a range of RQ values as noted above.
In both applications, results can be affected by DNA quality and primer design, particularly for amplicons under 100 bp (Ma, 2015.) A schematic representation of the Taqman reaction can be seen to the left, courtesy of UPODMG 1516 irivbrro
ddPCR
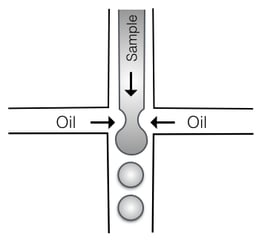 Droplet digital PCR (ddPCR) works by using oil droplets to partition a single sample into thousands of PCR reactions using Taqman chemistry (see image, left, of droplet immersion technique, courtesy of CAR15.) Each droplet contains a primer and probe set for both the gene of interest and the reference sequence. At the end of the run, each droplet is read as positive or negative for each reaction, math happens (this time, Poisson statistics), and the readout gives an absolute value of DNA concentration. By comparing the number of positive droplets for the gene of interest and reference gene, copy number can be determined through concentration values, rather than by comparison to a standard curve, and most results can be confidently interpreted as integers. Since each sample is partitioned into thousands of reactions, it does not need to be run in triplicates.
Droplet digital PCR (ddPCR) works by using oil droplets to partition a single sample into thousands of PCR reactions using Taqman chemistry (see image, left, of droplet immersion technique, courtesy of CAR15.) Each droplet contains a primer and probe set for both the gene of interest and the reference sequence. At the end of the run, each droplet is read as positive or negative for each reaction, math happens (this time, Poisson statistics), and the readout gives an absolute value of DNA concentration. By comparing the number of positive droplets for the gene of interest and reference gene, copy number can be determined through concentration values, rather than by comparison to a standard curve, and most results can be confidently interpreted as integers. Since each sample is partitioned into thousands of reactions, it does not need to be run in triplicates.
ddPCR is an extremely accurate application, even in areas of the genome that are difficult to accurately read (Bell, 2018.) Even though it does not require a standard curve or a normal control, it can take a few hours longer than qPCR to run, as the method is dependent on number of samples rather than number of cycles.
Choosing a method
As with all experiments, a number of factors will determine which approach is right for your experiment. You’ll have to consider:
- Resources needed
- Most molecular biology labs and departments will have qPCR machines available for use. ddPCR machines are not as commonly available and may not be an option to all researchers.
- Accuracy needed
- Do you need to call exact copy numbers or just general trends? How confident do you need to be in your determination to move forward with your experiment?
- Complexity of the gene of interest
- Some gene sequences are harder to amplify and quantify than others. If available, ddPCR may be a better option for more challenging cases.
- Price point
- If budget is a concern, look for the lowest-cost option that provides you confidence in the results you need.
Whichever method you choose, resist the temptation to over interpret your results: Sybr cannot be used to determine exact copy number and it may not be able to identify trends in cases where the copy number variation is two or less (Muhamad, 2011). Ambiguous Taqman or ddPCR results need to be rerun in order to be called accurately, and raw data should be checked to confirm negative samples and, if unexpected, verified by running the product on a gel. And only ddPCR provides absolute quantitative data, instead of relative quantification.
Even though no one option is perfect, with all this variance in your variant options, you’re likely to find an application that matches your needs. Happy quantifying!
References and Resources
More Resources on the Addgene blog
Droplet Digital PCR for AAV Quantification
Polymerase Chain Reaction Overview and Applications
Resources on Addgene.org
- Protocol: Polymerase Chain Reaction (PCR)
- Protocol: How to Design a primer
- Molecular Biology Reference
- Molecular Cloning Techniques
References
Bell AD, Usher CL, McCarroll SA. Analyzing Copy Number Variation with Droplet Digital PCR. Methods Mol Biol. 2018;1768:143-160. doi: 10.1007/978-1-4939-7778-9_9. PMID: 29717442.
Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005 Mar;5(2):209-19. doi: 10.1586/14737159.5.2.209. PMID: 15833050.
Ma L, Chung WK. Quantitative analysis of copy number variants based on real-time LightCycler PCR. Curr Protoc Hum Genet. 2014;80:7.21.1-7.21.. Published 2014 Jan 21. doi:10.1002/0471142905.hg0721s80
Colborn JM, Byrd BD, Koita OA, Krogstad DJ. Estimation of copy number using SYBR Green: confounding by AT-rich DNA and by variation in amplicon length. Am J Trop Med Hyg. 2008 Dec;79(6):887-92. PMID: 19052298.
Muhamad P, Chaijaroenkul W, Congpuong K, Na-Bangchang K. SYBR Green I and TaqMan quantitative real-time polymerase chain reaction methods for the determination of amplification of Plasmodium falciparum multidrug resistance-1 gene (pfmdr1). J Parasitol. 2011 Oct;97(5):939-42. doi: 10.1645/GE-2792.1. Epub 2011 May 9. PMID: 21554069.
Haridan, U. S., Mokhtar, U., Machado, L. R., Abdul Aziz, A. T., Shueb, R. H., Zaid, M., Sim, B., Mustafa, M., Nik Yusof, N. K., Lee, C. K., Abu Bakar, S., AbuBakar, S., Hollox, E. J., & Boon Peng, H. (2015). A comparison of assays for accurate copy number measurement of the low-affinity Fc gamma receptor genes FCGR3A and FCGR3B. PloS one, 10(1), e0116791. https://doi.org/10.1371/journal.pone.0116791
Providenti MA, O'Brien JM, Ewing RJ, Paterson ES, Smith ML. The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J Microbiol Methods. 2006 Jun;65(3):476-87. doi: 10.1016/j.mimet.2005.09.007. Epub 2005 Oct 10. PMID: 16216354
Topics: PCR





Leave a Comment