For the last 14 years, scientists have been coming to the Addgene website in search of plasmids. Now, scientists are beginning to see Addgene as a large data set. Addgene has over 65,000 plasmids in the repository, each verified by sequencing, which makes the repository a convenient source of sequence data.
A group of scientists from MIT tapped into this data to learn about trends in synthetic biology and DNA synthesis. They published their results in a paper in Nature Communications announcing a new bioinformatics tool that can predict whether a gene is natural or synthetic just by looking at its sequence.
MIT collaboration for gene classification
This inquiry started when Neil Thompson, a research scientist at MIT’s Computer Science and Artificial Intelligence Lab and a Visiting Professor at the Lab for Innovation Science at Harvard, wanted to see how new tools drive innovation in synthetic biology. “Synthetic biology, in particular, the ability to do DNA synthesis, is evolving so fast that it is a really nice opportunity to study it and see what happens when technological change happens really fast,” Thompson says.
To do this, Thompson reached out to MIT labs in the Synthetic Biology Engineering Research Consortium. That is how Aditya Kunjapur, a MIT graduate student at the time, got involved. Along with Philipp Pfingstag, a visiting graduate student from TU München at the time, the team built an algorithm that predicts whether a gene is synthetic or natural with 97.7% accuracy.
Classifying genes as natural or synthetic
But how can you tell if a gene is synthetic or natural? The answer to this lies in the codon usage biases of different organisms. Different species have different preferences for the codons they use; even if two species encode the same protein, their gene sequence may have slight or large variations depending on how closely related these species are. If a gene was moved exactly as is from one organism to a distantly related organism, the gene may not express well because of these different codon usage biases. But if a gene’s sequence was “optimized” so that it more closely resembles that of the host organism, it may express protein more efficiently.
To classify gene sequences as natural or synthetic, the researchers looked at intrinsic properties (GC content or rare codon percentage) and comparative properties (percent sequence identity and percent query coverage) that could be determined through comparisons with a reference sequence database. Through a series of simulations and machine learning, they created a classifier that predicts whether a gene was natural or synthetic 97.7% of the time in a test set of 173 known synthetic and natural genes.
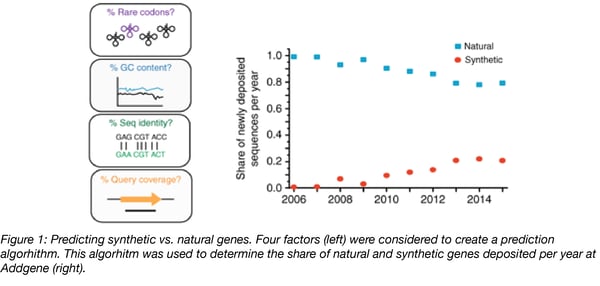
In designing and optimizing their algorithm, they found that GC content and rare codon percentage were not good predictors. There is not much variation in GC content between many organisms and DNA synthesis companies often work within a narrow range for GC content. Rare codons also are not good predictors because they are just that: rare. “Nature has many genes where there are not rare codons and many genes where there are instances of rare codons, sometimes intentionally modulating expression in ways we don’t quite understand yet,” Kunjapur says. Percent sequence identity was a different story - the team found that the percent sequence identity between the natural gene and a synthetic version of that gene was a clear predictor of whether the gene is synthetic or natural. They saw that a percent sequence identity below 85% correlates with synthetic sequences.
The researchers then used their classifier to identify synthetic and natural sequences in Addgene’s database. They showed a growing trend of sourcing genes from distantly related organisms. In 2006, less than 1% of the genes deposited at Addgene were synthetic. But nine years later, over 20% of the genes deposited that year were synthetic. They also found out that the most common expression system within the Addgene repository is mammalian, but the largest source of unique gene sequences is Proteobacteria - the most frequent transfer of genes is from Proteobacteria to mammalian expression vectors. And more broadly, their data revealed that the longer a natural gene sequence is, the less likely it is transferred into another organism in the lab.
Predicting lab-of-origin of engineered DNA
This isn’t the only time researchers have used Addgene as a large data set. Alec A. K. Nielsen & Christopher A. Voigt also recently published a tool (trained on Addgene plasmid datasets) to predict the lab-of-origin of engineered DNA. Both of these groups have generated robust tools that have practical applications in monitoring the accidental or intentional release of engineered organisms. “Is [the organism] engineered? Where did it come from? What does it do? Those are the sets of questions one would want to have answered with a biosurveillance tool,” Kunjapur says. Predicting lab-of-origin and synthetic vs. natural gene are essential and complementary tools in a time where biological engineering and synthetic biology is expanding rapidly.
References
- Read blog posts about synthetic biology
- Learn more about codon usage biases
- Check out Addgene's synthetic biology page
- Search for a plasmid
- Find Addgene's most popular plasmids
Topics: Synthetic Biology, Other





Leave a Comment