High-throughput cloning, in a nutshell, is the systematic combination of different genetic sequences into plasmid DNA. In high throughput cloning techniques, although the specific sequences of the genetic elements may differ (e.g., a set of various mammalian promoters), the same cloning procedure can be used to incorporate each element into the final construct. This strategy can be used to build vectors with diverse functions, and thus, is used in many biological fields. In synthetic biology for example, high-throughput cloning can be used to combine the functions of different genetic elements to generate non-natural tools such as novel biological circuits or sensors. Given the expanding palette of fluorescent proteins and the availability of powerful imaging technologies, the combination of multiple fluorescent protein sequences to develop diverse fluorescent reporters is a useful application of high-throughput cloning. MXS Chaining is one such technique and has been used to produce complex fluorescent reporter constructs. These fluorescent reporters can be used to detect structure and protein localization, as well as cellular processes like gene expression and cell migration (Sladitschek and Neveu, 2015).
Origin and purpose of MXS chaining
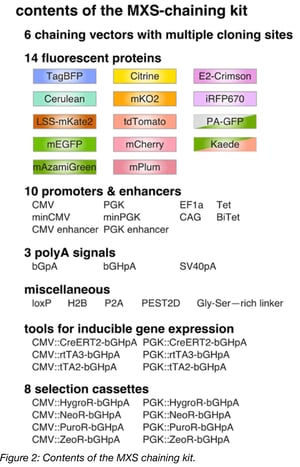
MXS-chaining was designed to create plasmids for fluorescence imaging or flow cytometry applications in mammalian cells. The modules used in this method include fluorescent proteins, promoters, enhancers, polyadenylation signals, inducible gene-expression sequences, and more. By combining these components, one can generate constructs to visualize cell cycle dynamics, titrate inducible transgene expression, or a variety of other clever applications.
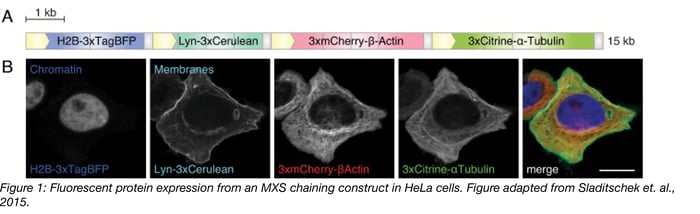
In one example, modular constructs generated with MXS-chaining were used to label subcellular structures in live cells. Four individual expression constructs were created, each containing a unique fluorescent protein with minimal spectral overlap between them (Table 1). In each construct, 3 copies of the fluorescent protein were fused in-frame to a tethering partner, which directed localization of the fluorophore, and thus enabled labeling of particular structures (Table 1). Each construct was flanked with a CMV promoter (to drive high-level expression) and a polyA signal (to terminate transcription) and the four constructs were then combined to create a single polycistronic 15 kb insert. The final construct was then introduced to HeLa cells (Figure 1). The resulting HeLa cells show robust labeling, with strong expression and detection of each fluorophore at the respective subcellular structure (Figure 1). Here, MXS Chaining gave researchers the ability to generate the four initial unique constructs and later combine all four constructs into one 15 kb insert.
Table 1: MXS Constructs for Labeling Cellular Structures
| Construct | Fluorophore | Excitation/emission maxima | Tethering partner | Subcellular localization |
| 1 | TagBFP | 399nm/ 456nm | histone 2B (H2B) | Chromatin |
| 2 | Cerulean | 433nm/ 475nm | Lyn-tag (derived from the tyrosine-protein kinase Lyn) | Membranes |
| 3 | mCherry | 587nm/ 610 nm | human β-Actin | Actin |
| 4 | Citrine | 516nm/ 529nm | human α-Tubulin | Tubulin |
Repeatable, directional cloning
The principle behind MXS-chaining is repeatable, restriction-enzyme based (chaining-based) cloning. The modules (Figure 2) are contained in individual plasmids and are each flanked by the same multiple cloning site (MCS). Modules are assembled one at a time by standard restriction enzyme digestion followed by ligation. Upon ligation, the original pattern of the MCS is regenerated at the flanks of each ligation product, which is the basis for the repeatability of the method. Note that the restriction sites of the MCS are only regenerated at the 5’ and 3’ termini of the ligation product, and the restriction site between the modules is eliminated upon ligation. As a result, once two modules are assembled next to each other, a third module cannot be placed between them, but rather, can only be added to the 5’ or 3’ terminus using the newly generated MCS. MXS-chaining also has the advantage of being directional, meaning that (in the previous example) one can control whether the third module is added to the 5’ terminus or the 3’ terminus of the ligation product.
Comparison to other high throughput cloning strategies
There are several high-throughput cloning strategies available for assembling these modular types of constructs, and different methods are suited for particular downstream applications. The tradeoffs between various cloning methods are listed in Table 2. For example, chaining-based cloning methods require that the separate modules combined using these techniques are free of specific restriction enzyme recognition sites. Thus, chaining methods are not necessarily suitable for combining endogenous sequences.
Table 2: High Throughput Cloning Strategies
| Method | Technology | Pros | Cons | References |
| Golden Gate | Type II restriction enzymes | Can assemble up to 10 modules in a single step. Through iteration of this step, the final construct can contain an unlimited number of modules. | Constraints on the order of the modules, because neighboring fragments require compatible cohesive ends | Engler et al., 2008 |
| Gibson Cloning | Gibson assembly method | Single reaction, can assemble large DNA sequences, no restriction enzymes needed | Not optimal for joining sequences with a high degree of identity, so not suited to generate polycistronic containing multiple identical polyadenylation signals or promoters | Gibson et al., 2009 |
| MXS-Chaining, BioBricks, Bglbricks, etc. | Chaining-based methods (restriction enzyme-based). | Well suited for the modular assembly of highly similar or repetitive sequences. Intermediate chaining products (called “cassettes”) can be readily reused in the assembly of any later construct (i.e.,functional cassettes are recyclable) | Can only work if the restriction sites used for the assembly are not found within the modules themselves (so may not be suitable for endogenous coding sequences), so enzyme choice is critical. May not support in-frame fusions of coding sequences |
Sladitschek and Neveu, 2015; Shetty et al., 2008; Anderson et al., 2010 |
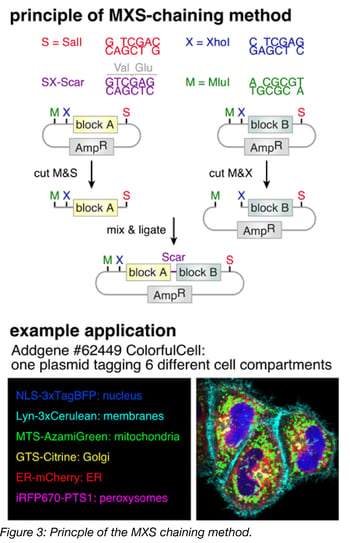 Among the chaining-based methods, the specific restriction enzymes used affect what general types of modules can be assembled. For example, because CpG dinucleotides are underrepresented in most vertebrate genomes (Josse et al., 1961, Swartz et al., 1962), SalI, XhoI and MluI recognition sites, which contain CpG dinucleotides in their 6-mer sequences, are rarely found in the transcriptomes of these species and can often be used to clone cDNAs from them. These enzymes might not, however, be appropriate for cloning genes from organisms with high CpG dinucleotide representation. The specific restriction enzymes used in various chaining-based cloning methods are listed in Table 3.
Among the chaining-based methods, the specific restriction enzymes used affect what general types of modules can be assembled. For example, because CpG dinucleotides are underrepresented in most vertebrate genomes (Josse et al., 1961, Swartz et al., 1962), SalI, XhoI and MluI recognition sites, which contain CpG dinucleotides in their 6-mer sequences, are rarely found in the transcriptomes of these species and can often be used to clone cDNAs from them. These enzymes might not, however, be appropriate for cloning genes from organisms with high CpG dinucleotide representation. The specific restriction enzymes used in various chaining-based cloning methods are listed in Table 3.
The goal of high throughput cloning is to facilitate the construction of plasmids with many components. The creators of the MXS-chaining method have used the technique to engineer various constructs with applications in flow cytometry approaches in mammalian cell culture systems (their original goal). Their work has demonstrated that the MXS-chaining method can be used to build these types of constructs robustly, quickly, and simply. We hope that you can similarly apply MXS chaining to your experimental needs. Let us know how you use MXS Chaining by emailing us at blog@addgene.org.
Table 3: Chaining Based Cloning Methods
| Method | Restriction Enzymes | |
| Chaining | Direction | |
| MXS-Chaining | SalI and XhoI | MluI |
| BioBricks | SpeI and XbaI | EcoRI and PstI |
| Bglbricks | BglII and BamHI | EcoRI |
Additional Resources on the Addgene Blog
- Read about Gibson Assembly
- Learn About Other Plasmid Cloning Techniques
- Plasmids 101: Green Fluorescent Protein
Resources on Addgene.org
- Browse our Fluorescent Protein Collection
- Find Plasmid Kits at Addgene
- Find Plasmid Cloning Protocols
References
1. Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, et al. BglBricks: A flexible standard for biological part assembly. Journal of biological engineering. 2010;4(1):1. Pubmed PMID: 20205762. PubMed Central PMCID: PMC2822740.
2. Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PloS one. 2008;3(11):e3647 doi: 10.1371/journal.pone.0003647. Pubmed PMID: 18985154. PubMed Central PMCID: PMC2574415.
3. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO.Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009. May;6(5):343–345. Pubmed PMID: 19363495.
4. Josse J, Kaiser AD, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. The Journal of biological chemistry. 1961. March;236:864–875. Pubmed PMID: 13790780.
5. Shetty RP, Endy D, Knight TF. Engineering BioBrick vectors from BioBrick parts. Journal of biological engineering. 2008;2:5. Pubmed PMID: 18410688. PubMed Central PMCID: PMC2373286.
6. Sladitschek HL, Neveu PA. MXS-Chaining: A Highly Efficient Cloning Platform for Imaging and Flow Cytometry Approaches in Mammalian Systems. PLoS One. 2015 Apr 24;10(4):e0124958. PubMed PMID: 25909630. PubMed Central PMCID: PMC4409215.
7. Swartz MN, Trautner TA, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. The Journal of biological chemistry. 1962. June;237:1961–1967. Pubmed PMID: 13918810.
Topics: Fluorescent Proteins, Generating Fusions







Leave a Comment