 Hodaka Fujii, M.D., Ph.D., is an Associate Professor at Osaka University. The Fujii lab specializes in developing novel technologies to analyze molecular mechanisms of genome functions such as epigenetic regulation and transcription by using locus-specific chromatin immunoprecipitation (locus-specific ChIP). These methods consist of insertional chromatin immunoprecipitation (iChIP) and engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP), both developed in the lab. In June 2014, Dr. Fujii joined Addgene's Advisory Board.
Hodaka Fujii, M.D., Ph.D., is an Associate Professor at Osaka University. The Fujii lab specializes in developing novel technologies to analyze molecular mechanisms of genome functions such as epigenetic regulation and transcription by using locus-specific chromatin immunoprecipitation (locus-specific ChIP). These methods consist of insertional chromatin immunoprecipitation (iChIP) and engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP), both developed in the lab. In June 2014, Dr. Fujii joined Addgene's Advisory Board.
Addgene: Your lab has worked extensively with enChIP systems. Can you describe this technology and its advantages?
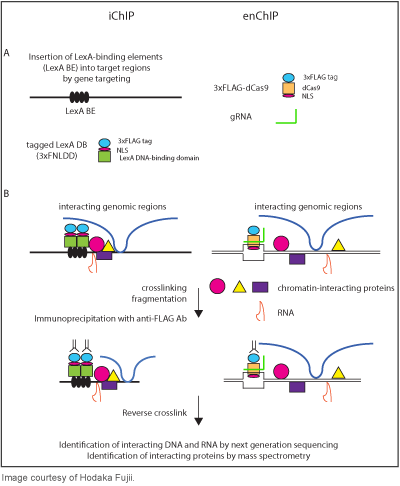
Fujii: In the last several years, my lab has been working on development of technologies for biochemical analysis of genome functions such as transcription and epigenetic regulation. To elucidate molecular mechanisms of regulation of genome functions, we need to identify molecules associated with specific genomic regions of interest in a non-biased manner. To achieve this goal, it is necessary to isolate specific genomic regions while retaining molecular interactions. For this purpose, we developed the insertional chromatin immunoprecipitation (ChIP) (iChIP) technology, which uses locus-tagging and affinity purification, in 2009. However, iChIP requires insertion of recognition sequences of an exogenous DNA-binding protein such as LexA into the target locus, which is the rate-limiting step. When engineered DNA-binding molecule technologies such as TAL proteins and CRISPR became available, I was very excited because genome-editing with these molecules makes it much easier to knock-in LexA binding elements into the target region. However, it struck me that direct use of such engineered DNA-binding molecules even enables us to skip the step of knocking-in. In 2013, we showed that isolation of a single copy genomic locus is feasible by using those engineered DNA-binding molecules for the first time and named the new technology as engineered DNA-binding molecule-mediated ChIP (enChIP). By using enChIP, specific genomic regions can be easily isolated for identification of associated proteins, RNAs, and other genomic regions. I believe that enChIP will markedly accelerate research of regulation mechanisms of genome functions in the hope of developing drugs against intractable diseases. Currently, a number of labs are using enChIP, and I'm very happy to see that many researchers all over the world have requested plasmids for enChIP through Addgene.
Retroviral Expression System for enChIP using CRISPR
Addgene: Your lab recently developed a retroviral enChIP system using CRISPR technology. Can you describe this technology and some of its potential applications?
Fujii: We generated a series of retroviral vectors expressing tagged dCas9 and guide RNA. The retroviral enChIP system enables us to express the CRISPR complex much easier in various cells, including primary mouse cells. We believe that this system will allow researchers to use enChIP in a wide variety of biological systems. We are currently developing several additional systems of enChIP to make the technology more user-friendly and precise. We will deposit the plasmids of these systems to Addgene in the near future.
Addgene: Has your lab been working on any other technologies that you would like to describe?
Fujii: I believe that "Biology is Method", i.e. new technologies often lead to big discoveries in biology. Therefore, my lab is dedicated to development of new biological technologies. The locus-specific ChIP technologies consisting of iChIP and enChIP are recent examples. We also developed the inducible translocation trap (ITT) system to analyze signal-induced nuclear translocation of proteins in 2004. In addition, we have recently developed the oligoribonucleotide (ORN) interference-PCR (ORNi-PCR) method to suppress PCR amplification of specific DNA sequences. Thus, we've made some contributions in technology development in biology and hope to develop more in the future.
Involvement in the Scientific Community
Addgene: In 2012 your lab first deposited plasmids with Addgene. In 2014, you joined Addgene's Advisory Board, as well as the Editorial Board of Scientific Reports and PLoS One. Additionally, you are involved in multiple professional scientific societies. Why is being involved in the scientific community important to you and your lab?
Fujii: I first knew Addgene around 2010 and found the scheme very nice at first sight in that Addgene takes care of everything including MTA and shipping. Addgene realizes the One-Stop shopping for a wide range of plasmids. The merit of depositing plasmids to Addgene has been spectacular. We've got more than one hundred requests, and it would have been very difficult for us to respond to such a large number of requests by ourselves without Addgene. So, when I received an invitation to join the Advisory Board of Addgene, I accepted the offer without any hesitation. I hope I will be able to effectively promote Addgene, which is also beneficial to my research.
I believe that involvement in the scientific community is a kind of duty for researchers when considering the peer-review system of journals and grant applications, for example. However, at the same time, it's fun to be involved in these professional activities because I can contribute by, for example, helping to improve the papers submitted to journals. In addition, it is my pleasure to interact with my colleagues in meetings of scientific societies.
Addgene: Throughout your career, you have held research appointments in the US (New York University School of Medicine), Europe (Basel Institute for Immunology), and Japan (University of Tokyo, Osaka University). Can you speak about your different experiences in research labs. What are some similarities and differences you have observed as you have traveled the world?
Fujii: Fortunately, I was able to work in different parts of the world in my early research career. I've found that research institutes in different places have very different cultures. In Basel, I felt that the time flows much slower, like the flow of the Rhine River. In the US, I very much enjoyed interactions with many excellent colleagues at NYU and visiting scientists, such as seminar speakers. They made an every effort to interact with each other, which is critically important, especially for young researchers. It was a really nice incubator for me. In Japan, of course, it is optimized for living for Japanese, which is not trivial, and the grant situation is currently much better than that in the US for me. In addition, I've been very lucky to work with excellent colleagues in my lab. On the other hand, I feel that researchers in Japan are not eager to interact, which I don't think is good for Japanese science. I really miss the intellectual environment in the US in this regard. In addition, not many researchers in Japan aim to develop new technologies. Majority of them are eager to just "import" technologies developed abroad, so it's a little boring. I think that such tendency is one of the reasons why we have only three biology-related Nobel prizes, out of which only one was done in Japan, which is very disappointing. I hope I can help change the situation in the future.
Addgene: What advice do you give to post-docs and graduate students as they complete their appointments in your lab?
Fujii: These days it is very difficult to survive as a researcher because of the tough competition for academic positions and poor grant situation. So, young researchers need to publish good publications in a short period of time and may need to do some fashionable projects. However, I hope that they also have long-term goals and continue to do research to realize their vision. Their progress might be slow, but if they don't stop, they may be able to achieve something that may change the rules of the game. In this regard, I believe that development of new technologies may be very effective in transforming conventional research. So, I hope my younger colleagues may be able to develop new technologies, hopefully they'll be completely different from what we are now doing in my lab then when they become independent.
Looking for enChIP Plasmids?
- All Hodaka Fujii lab plasmids
- Sign up to receive email alerts when new plasmids from the Fujii lab become available. Visit Hodaka Fujii lab's Addgene page and click the "Subscribe to Alerts" link in the Addgene Alerts box.
References
- Identification of Proteins Associated with an IFNgamma-Responsive Promoter by a Retroviral Expression System for enChIP Using CRISPR. Fujita T, Fujii H. PLoS One. 2014 Jul 22;9(7):e103084. doi: 10.1371/journal.pone.0103084.
- Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Sci Rep. 2013 Nov 8;3:3171. doi: 10.1038/srep03171.
- Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR.
Fujita T, Fujii H. Biochem Biophys Res Commun. 2013 Aug 11. pii: S0006-291X(13)01329-6. doi: 10.1016/j.bbrc.2013.08.013.
Topics: CRISPR, CRISPR Expression Systems and Delivery Methods






Leave a Comment