This post was contributed by guest blogger, Leo Vo, a PhD candidate in the Sternberg Lab at Columbia University Medical Center.
DNA transposons are ubiquitous genetic elements capable of spreading within and between genomes, and have been adapted for a wide variety of applications - as mutagenesis tools for genetic studies (e.g. using mariner), as a method of tagmentation for next-gen sequencing pipelines (e.g. using Tn5), and as knock-in tools for human therapeutics (e.g. using Sleeping Beauty). Despite their diverse utility, DNA transposons have been hindered by their limited programmability: depending on the transposases involved, they insert either randomly, or at fixed genomic target sites.
This obstacle was recently surpassed with the discovery of CRISPR-transposons by the Sternberg and Zhang labs (Peters et al., 2017; Klompe et al., 2019; Strecker et al., 2019), which direct RNA-guided transposition. By natively combining the DNA integration capabilities of transposases and the target programmability of CRISPR-Cas, these systems represent the next generation of tools for efficient and programmable insertions of large DNA cargos into bacterial genomes. This new capability was named INTEGRATE (Insertion of Transposable Elements by Guide RNA-Assisted Targeting) by the Sternberg group, and CAST (CRISPR-associated Transposase) by the Zhang group.
Mechanism of CRISPR-transposons
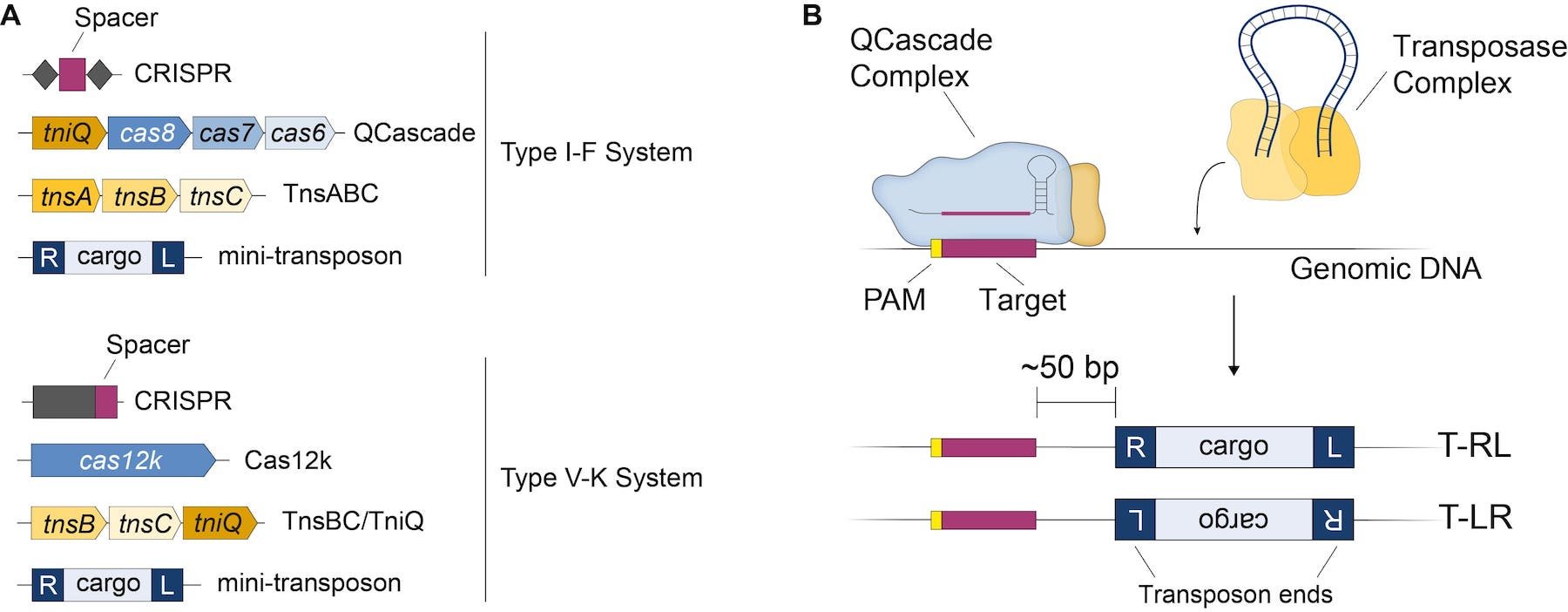
One of the CRISPR-transposon systems developed by the Sternberg lab derives from Vibrio cholerae (V. cholerae INTEGRATE - hereafter VchINT) and encodes a Type I-F CRISPR-Cas system. The four major components required for RNA-guided DNA integration are (Fig. 1A, top):
- A CRISPR RNA (crRNA) comprising a 32 nt spacer sequence specifying the target site.
- Four proteins (TniQ, Cas8, Cas7, Cas6) that forms the QCascade DNA-targeting module together with the crRNA.
- Three transposase proteins (TnsA, TnsB, TnsC).
- The donor DNA (aka mini-transposon), which contains the DNA cargo of interest flanked by ~50-150 bp transposon end sequences (R and L ends).
|
|
|
Figure 1: (A) Components involved in each type of CRISPR-Transposon system. (B) Mechanism of RNA-guided transposition in type I-F INTEGRATE. |
The requirement for multiple Cas proteins to form the Cascade targeting complex is a hallmark of Type I CRISPR-Cas systems, in contrast to single-protein effectors employed by Type II systems (such as CRISPR-Cas9). Unlike in regular Type I CRISPR-Cas systems, however, Cascade from VchINT does not recruit other nucleases (such as Cas3) or have its own nuclease activity. Instead, it forms a novel co-complex with the transposition protein TniQ for targeting (Halpin-Healy et al., 2020). Genomic targets require an upstream 2-bp PAM sequence for recognition by QCascade; although 5’-CC-3’ is one of the best PAMs for VchINT, many other PAMs are also efficiently recognized by the system.
Guided by the crRNA, QCascade complexes survey the genome and bind target DNA sequences using RNA-DNA base pairing. Separately, TnsABC forms a transposase complex with the mini-transposon, excises it from the donor molecule, and inserts it ~50 bp downstream of the genomic target site bound by QCascade (Fig. 1B). The process also creates a 5-bp target-site duplication (TSD), a hallmark characteristic of the Tn7-family of transposons. Insertion of the mini-transposon can occur in two different orientations, but is strongly biased towards T-RL, where the R end is proximal to the target site (Klompe et al., 2019, Vo et al., 2020).
Type V-K CRISPR-transposon systems function in a very similar manner to Type I-F, but instead employ a Cas12k protein instead of three Cas proteins (Fig. 1A, bottom), and require only two transposase proteins (TnsB and C). While requiring fewer components for RNA-guided DNA integration, Type V-K systems frequently produce off-target DNA insertions and intermediate structures known as cointegrates (Vo et al., 2020; Strecker et al., 2020).
CRISPR-transposons for bacterial genomes engineering
In the Sternberg group’s initial report of VchINT, three separate plasmids and multiple promoters were used to express the necessary components in E. coli (Klompe et al., 2019). Recently, the lab introduced a streamlined expression approach (Fig. 2A), involving two-plasmid or single-plasmid systems (Vo et al., 2020). In the two-plasmid system, an effector plasmid (pEffector) expresses all seven required genes and the CRISPR RNA under the same promoter, and a donor plasmid (pDonor) encodes the mini-transposon. pEffector and pDonor can also be combined in single-plasmid INTEGRATE (pSPIN), which offer simple transformations for smaller cargos, and the ability to easily cure cells of the plasmid after integration.
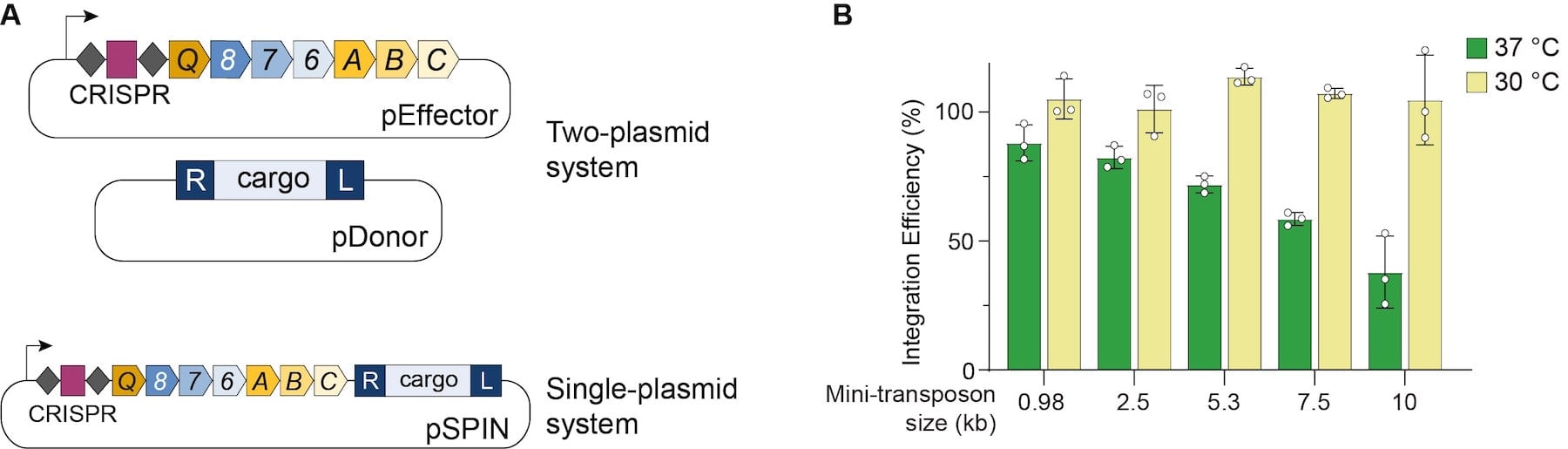 |
| Figure 2: (A) Streamlined VchINT expression constructs. (B) High integration efficiencies achieved with VchINT, at two different incubation temperatures. Data adapted from Vo et al., 2020. |
In E. coli, the new streamlined VchINT system was shown to catalyze insertion of mini-transposons up to 10 kb in size, with efficiencies of 90-100% (Fig. 2B), and >99% on-target specificity. Additionally, by expressing multiple crRNAs in parallel (Fig. 3), multiplexed insertions into multiple distinct targets could be achieved (Vo et al., 2020). VchINT was also shown to be active in various gram-negative bacterial hosts of interest, including within mixed communities and into species not previously cultured or isolated (Rubin et al., 2020).
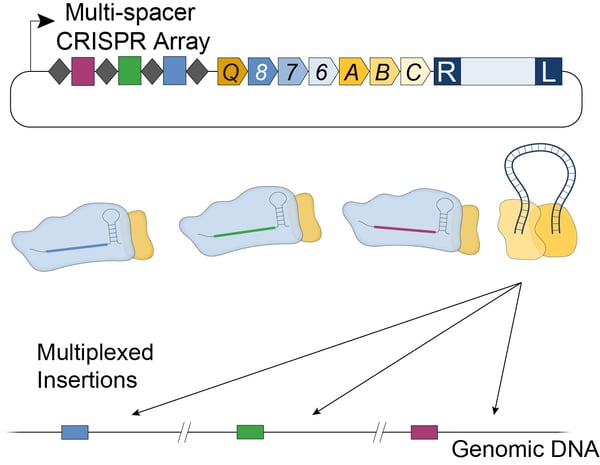 |
| Figure 3: Multiplexed insertions with Type I-F INTEGRATE using a single plasmid containing a multi-spacer CRISPR array. |
Workflow and technical considerations
Making insertions in bacterial genomes with the V. cholera INTEGRATE system follows a simple workflow:
- Clone spacer(s) into the CRISPR array
- Clone desired DNA cargo into the mini-transposon
- Transform plasmid(s) into cells and plate overnight
- Screen colonies phenotypically or by PCR/sequencing
While CRISPR-transposons are powerful and simple to use, users should keep aware of a few important details. For transposons to be specifically recognized during transposition, genetic cargos must be flanked by the L and R ends - thus, scarless insertions are not currently possible, unlike methods involving homologous recombination. Additionally, these transposon also exhibit something called target immunity, whereby multiple proximal insertions with the same transposon exhibit reduced efficiencies (Vo et al., 2020).
CRISPR-transposons within the CRISPR toolbox
RNA-guided transposases are exciting new additions to the growing CRISPR toolbox for genome engineering in bacteria, alongside more established tools such as Cas9-recombineering, CRISPRa/i, and base editing. Leveraging a unique marriage between distinct biological elements, CRISPR-transposons excel at programmable insertions of even very large genetic cargo, at one or multiple desired genomic loci. Many applications of these systems have already been demonstrated, ranging from large genomic insertions and/or deletions, to multiplexed gene knockouts (Vo et al., 2020), to multi-copy genomic integration of metabolic cassettes (Zhang et al. 2020); however, the technology potential enabled by CRISPR-transposons has yet to be fully realized.
 Many thanks to our guest blogger Leo Vo!
Many thanks to our guest blogger Leo Vo!
Leo Vo is currently a PhD candidate in the Sternberg Lab at Columbia University Medical Center. He currently studies CRISPR biology and CRISPR genome engineering technologies. Message or follow him on twitter @Vopinator.
References:
Halpin-Healy TS, Klompe SE, Sternberg SH, Fernández IS (2020) Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577:271–274 . https://doi.org/10.1038/s41586-019-1849-0
Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH (2019) Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571:219–225 . https://doi.org/10.1038/s41586-019-1323-z
Peters JE, Makarova KS, Shmakov S, Koonin EV (2017) Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci USA 114:E7358–E7366 . https://doi.org/10.1073/pnas.1709035114
Rubin BE, Diamond S, Cress BF, Crits-Christoph A, He C , Xu M, Zhou Z, Smock DC, Tang K, Owens TK, Krishnappa N, Sachdeva R, Deutschbauer AM, Banfield JF, Doudna JA (2020) Targeted Genome Editing of Bacteria Within Microbial Communities. bioRxiv https://www.biorxiv.org/content/10.1101/2020.07.17.209189v2
Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F (2019) RNA-guided DNA insertion with CRISPR-associated transposases. Science 365:48–53 . https://doi.org/10.1126/science.aax9181
Strecker J, Ladha A, Makarova KS, Koonin EV, Zhang F (2020) Response to Comment on “RNA-guided DNA insertion with CRISPR-associated transposases.” Science 368:eabb2920 . https://doi.org/10.1126/science.abb2920
Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH, Sternberg SH (2020) CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat Biotechnol. https://doi.org/10.1038/s41587-020-00745-y
Zhang Y, Sun X, Wang Q, Xu J, Dong F, Yang S, Yang J, Zhang Z, Qian Y, Chen J, Zhang J, Liu Y, Tao R, Jiang Y, Yang J, Yang S (2020) Multicopy Chromosomal Integration Using CRISPR-Associated Transposases. ACS Synth Biol 9:1998–2008 . https://doi.org/10.1021/acssynbio.0c00073
Additional resources on the Addgene blog
- Browse all of our CRISPR blog posts
- Check out other CRISPR methods for bacteria
Resources on Addgene.org
- Visit the CRISPR guide
- Find CRISPR plasmids for bacteria
Topics: CRISPR, Microbiology, Other CRISPR Tools






Leave a Comment