Every few months, we highlight a subset of the new plasmids, antibodies, and viral preps in the repository through our Hot Plasmids articles. Today, it's time for another edition of Hot Plasmids!
Here's what you'll find in this post:
- New compact, efficient prime editors
- A versatile and precise system to improve on-target delivery of genetic cargo
- Optogenetic proximity labeling and TransitID
- Tracking cellular history with protein ticker tape
- Teaching an old antibody new tricks
By Susanna Stroik
The Liu lab recently used phage-assisted continuous evolution and protein engineering to generate seven new prime editors, PE6a–g, that overcome some current prime limitations and enable impressive editing efficiencies in vitro and in vivo.
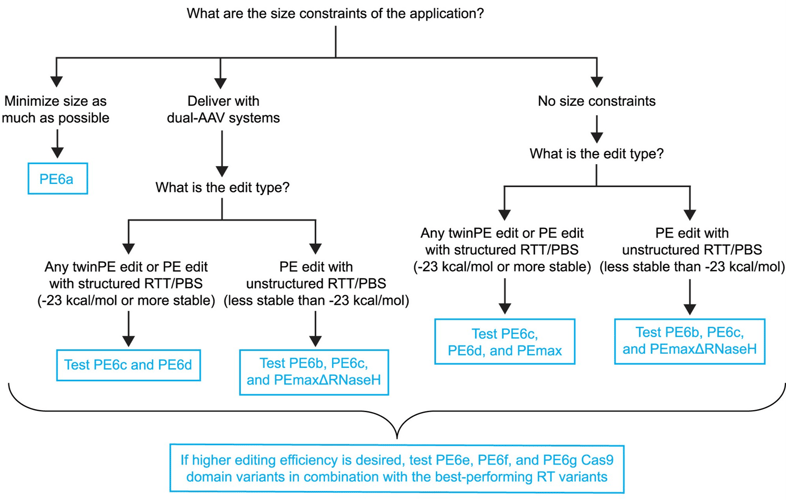
Incorporating reverse transcriptase (RT) variants, PE6a and PE6b are 500–800 bp smaller than previous editors, but just as effective and with no additional off-target edits, making them ideal for size-limited AAV delivery. PE6c and PE6d are optimized for dual-AAV systems. These variants use different RT enzymes and performed large edits with similar or better efficiency than PEmax, the current state-of-the-art. PE6b was very efficient for short, unstructured templates and significantly reduced undesired edits (indels, etc.), while PE6c and PE6d were most efficient at complex edits, such as long or highly structured templates. Cas9 variants PE6e–g often had even higher editing efficiencies but varied depending on the specific edit.
 |
|
Figure 1: Choosing the best PE6 variant for your application. Image reused form Doman and Pandey et al., 2023 under CC-BY 4.0 License. |
Doman, J.L., Pandey, S., et al. (2023). Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell, 186(18), 3983–4002.e26. doi: 10.1016/j.cell.2023.07.039. PMID: 37657419.
A versatile and precise system to improve on-target delivery of genetic cargo
By Alyssa Neuhaus
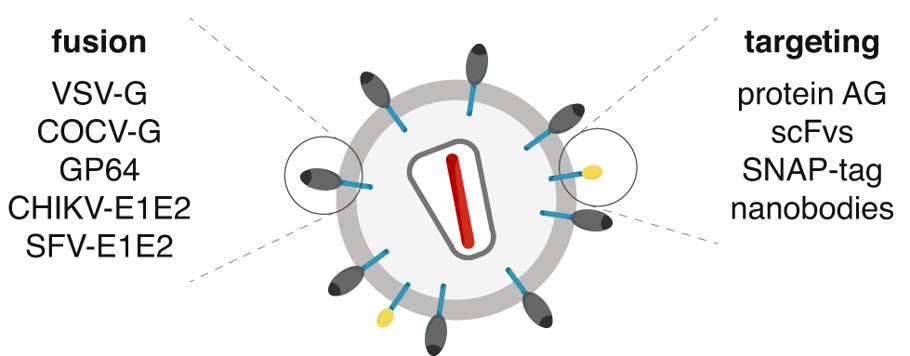
To improve delivery and reduce off-target effects of gene therapy, Feng Zhang’s lab has developed a system called DIRECTED (Delivery to Intended REcipient Cells Through Envelope Design). This modular approach combines an interchangeable set of fusogenic and targeting components to reprogram delivery vehicles like lentiviral vectors, CreVLPs, and eVLPs for in vivo and in vitro applications.
|
| Figure 2: Modular features of DIRECTED particles where the fusion and targeting components are interchangeable. Image from Strebinger et al., 2023, reused under CC-BY 4.0 License. |
Importantly, even commercial antibodies can be tethered on the viral surface, expanding the range of targetable cell types. DIRECTED is compatible with fusogens from different viral families and can be used to target multiple delivery systems, such as lentivirus and MMLV gag, to specific cell types. It can also make CAR-T cell therapy more accessible by simplifying the production process and could even enable the creation of CAR-T cells directly within the body.
Overall, DIRECTED can increase on-target delivery with various delivery modalities and has the potential to reduce negative side effects. This platform offers a promising approach for experimental therapies and medical applications.
Strebinger, D., et al. (2023). Cell type-specific delivery by modular envelope design. Nature Communications, 14(1), 5141. https://doi.org/10.1038/s41467-023-40788-8. PMID 37612276.
Optogenetic proximity labeling and TransitID
By Mike Lacy
Proximity labeling can identify protein interactions or map a local proteome, but some applications have been challenging or inaccessible. The Ting lab recently made two deposits that increase the utility of this technique.
By engineering a light-sensitive LOV domain into the TurboID enzyme, Lee and Cheah et al. achieved precise control over when and where the enzyme is active, drastically reducing background labeling caused by the biotin naturally present in experimental systems. They demonstrated LOV-Turbo in neurons, in pulse-chase experiments, and more.
Next, Qin and Cheah et al. described TransitID, where two proximity labeling enzymes, TurboID and APEX2, are localized to different cellular compartments. They showed APEX2 can use an orthogonal substrate, alkyne-phenol, to label proteins with an alkyne group for click chemistry reactions instead of biotin — a valuable development in and of itself. After adding one enzyme’s substrate, then the other, a dual-enrichment and mass spectrometry workflow identifies proteins bearing both labels (which had therefore been trafficked between locations).
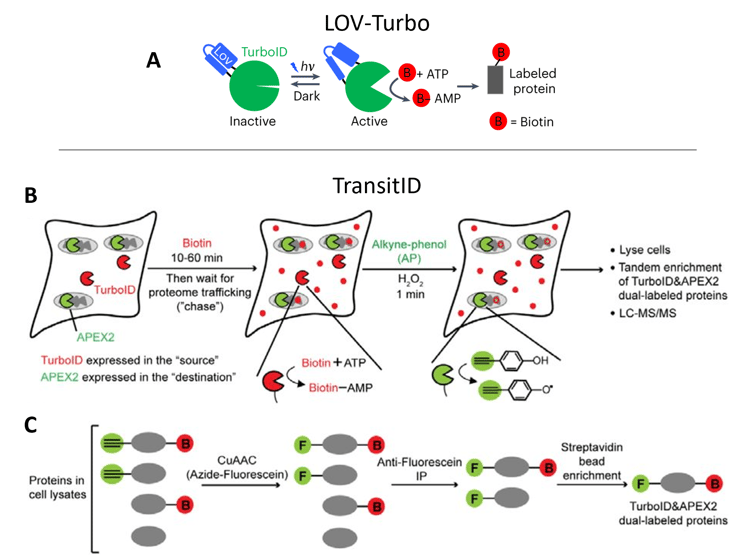 |
| Figure 3: A) LOV-Turbo becomes active when illuminated with blue light, catalyzing the biotinylation of nearby proteins. B) TransitID uses sequential labeling by TurboID and APEX2 at different cellular locations. C) Enrichment of dual-labeled proteins identifies proteins that had been trafficked from the “source” to “destination”. Images adapted from Lee and Cheah et al. 2023 bioRxiv (A) and Qin and Cheah et al. 2023 bioRxiv (B-C). |
They were able to label proteins moving to and from the nucleus, ER, mitochondria, stress granules, and more. For added precision, the authors also incorporated LOV-Turbo into TransitID experiments.
These two developments open up new possibilities for proximity labeling experiments in cells, tissues, and organisms.
Lee, S.Y., Cheah, J.S., et al. (2023). Engineered allostery in light-regulated LOV-Turbo enables precise spatiotemporal control of proximity labeling in living cells. Nature Methods, 20(6), 908–917. doi: 10.1038/s41592-023-01880-5. PMID: 37188954. Preprint available on bioRxiv – doi: 10.1101/2023.03.09.531939.
Qin, W., Cheah, J.S., et al. (2023). Dynamic mapping of proteome trafficking within and between living cells by TransitID. Cell, 186(15), 3307–3324.e30. doi: 10.1016/j.cell.2023.05.044. PMID: 37385249. Preprint available on bioRxiv – doi: 10.1101/2023.02.07.527548.
Tracking cellular history with protein ticker tape
By Brian O’Neill
The Cohen lab and colleagues recently created a system for recording the transcriptional history of a cell. Lin et al. generated a fusion protein called iPAK4 that assembles into a slow-growing protein fiber in the cytoplasm and can be fluorescently labeled. Using lentiviral vectors to stably or inducibly express tagged iPAK4 from different promoters, specific time points appear as fluorescent bands in the fiber, resembling a “ticker tape” recording.
The authors created externally-timed bands of color with HaloTag-iPAK4 by applying different HaloTag dyes and showed they could record transcriptional activation events as bands of induced GFP-iPAK4 expressed under promoters like the tet-responsive element (TRE) or the cFos promoter. For example, neurons treated with a phorbol ester activator gave fluorescent signal in the ticker tape from cFos::GFP-iPAK4 within an hour. Analyzing the ticker tape images could recover transcriptional histories, accurate to within 40 minutes.
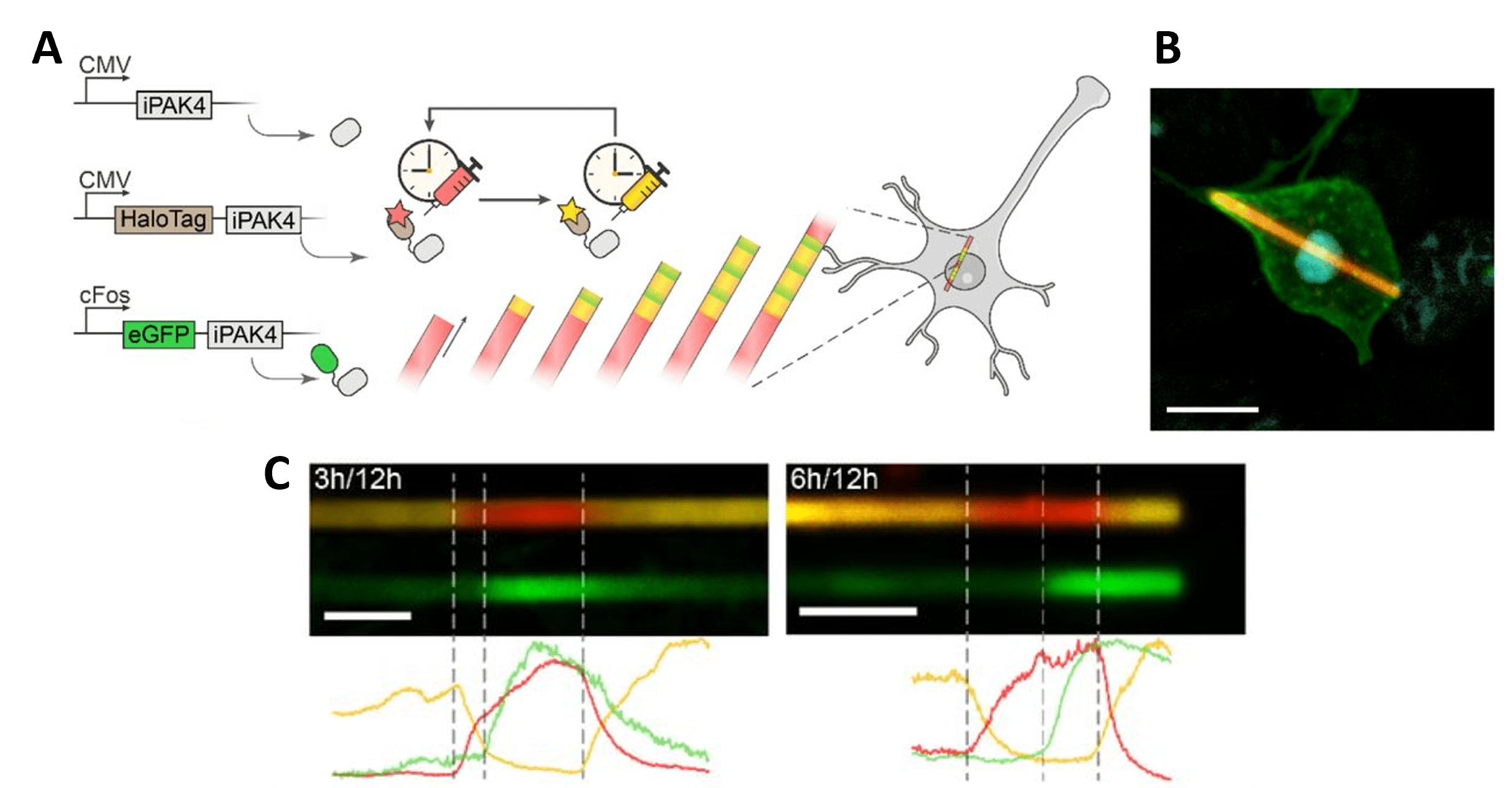 |
| Figure 4: Time-resolved protein ticker tape. A) Design of iPAK4 protein ticker tape recording. B) HEK cell with an iPAK4 fiber labeled with HaloTag-iPAK4. Scale bar 10 µm. C) Images of iPAK4 fibers in cultured rat hippocampal neurons. HaloTag dyes are switched from yellow to red at 0 h and back to red at 12 h, and PMA activation induces cFos::GFP-iPAK4 at 3 or 6 h. Scale bar 5 µm. Images adapted from Lin et al., 2023 bioRxiv, under CC-BY-NC 4.0 license. |
This system provides a unique new way to record and quantify the dynamics of promoter activity in single cells.
Lin D., Li X., et al. (2023). Time-tagged ticker tapes for intracellular recordings. Nature Biotechnology, 41(5), 631–639. doi: 10.1038/s41587-022-01524-7. PMID: 36593408. Preprint on bioRxiv — doi: 10.1101/2021.10.13.463862.
Teaching an old antibody new tricks
By Ashley Waldron
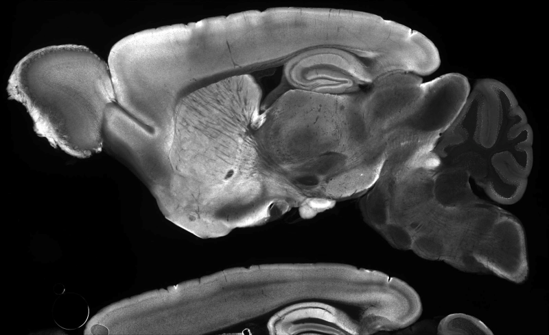
While IHC protocols typically call for thin sections of tissue, the Wu Lab is working to create a three-dimensional atlas of protein expression in the adult mouse brain using thick-section and whole-mount labeling techniques. The Trimmer lab’s anti-PSD-95 [K28/43R] recently passed their initial tests with thick slice (70 μm) immunohistochemistry (IHC) (Figure 5).
PSD-95 is a scaffolding protein known for its roles in synapse development, transmission, and plasticity and is a popular marker of excitatory synapses in assays like IHC. In fact, Anti-PSD-95 [K28/43R] has been one of the hottest antibodies in Addgene’s collection since we began distribution and has been verified to perform well in standard IHC and western blot protocols.
You can find more specific information on the Wu Lab’s use of Anti-PSD-95 [K28/43R] under the Application section of the Addgene antibody page, and learn more about the whole-brain reference atlas efforts, including a list of validated antibodies, on their webpage: mAb3D Atlas.
|
| Figure 5: Delipidated mouse brain tissue was stained with Anti-PSD-95 [K28/43R] (70 µm mounted sagittal section). Credit: Zhuhao Wu, Weill Cornell Medicine. |
Interested in this antibody, but wish it were from a different source species than mouse? No problem! We also have chimeric versions of Anti-PSD-95 [K28/43R] in rabbit, chicken, and rat.
Find Anti-PSD-95 antibodies here!
Andrews, N.P., et al. (2019). A toolbox of IgG subclass-switched recombinant monoclonal antibodies for enhanced multiplex immunolabeling of brain. eLife, 8, e43322. doi: 10.7554/eLife.43322. PMID: 30667360.
Topics: Hot Plasmids





Leave a Comment