This post was contributed by guest blogger, Krissy Lyon, a PhD candidate in Neuroscience at Harvard University.
 Just as computers, cell phones, and cars become more technologically advanced leaving earlier versions obsolete, the techniques we use in lab are replaced by improved versions that save both time and money. Yet, knowledge of historical techniques comes in handy whether you are perusing classic papers or are brainstorming new technological innovations. Let’s take a look at three historical techniques: southern blotting, restriction mapping, and sequencing gels, as well as their modern equivalents and see what we can learn from their evolution.
Just as computers, cell phones, and cars become more technologically advanced leaving earlier versions obsolete, the techniques we use in lab are replaced by improved versions that save both time and money. Yet, knowledge of historical techniques comes in handy whether you are perusing classic papers or are brainstorming new technological innovations. Let’s take a look at three historical techniques: southern blotting, restriction mapping, and sequencing gels, as well as their modern equivalents and see what we can learn from their evolution.

Southern blotting and quantitative real-time PCR
Southern blotting, named for its inventor Edward M. Southern (1), can be used to detect DNA identity, size, and abundance. If a scientist wants to know if an organism has a particular gene mutation, southern blotting can be used to identify the presence of that mutated gene. In southern blotting, DNA is fragmented into smaller pieces, run on a gel, and transferred to a membrane for analysis through hybridization of a radioactively or fluorescently labeled probe. If the DNA is complementary to the probe (for example, has the sequence of the mutated gene the researcher is searching for), then the probe will bind to the sample thereby labeling it. The label can then be visualized by autoradiography and X-ray film for radioactively labeled probes or by the presence of fluorescence for fluorescently labeled probes. Colorimetric and luminescence based detection methods are also commonly used. In all cases, the result can be quantified by the strength of the signal in comparison to a control sample.
Southern blotting applications range from confirming the results of cloning or knockout experiments to forensics and clinical diagnostics. One example of southern blotting being used to identify a gene mutation is the genetic test used for sickle cell anemia (2). Southern blotting, in conjunction with restriction digest analysis, is used to identify the presence of the mutated β globin gene that causes sickle cell anemia. The mutation happens to destroy a sequence recognized by the restriction enzyme MstII. When a restriction digest with MstII and southern blotting are conducted on a sample from an individual without sickle cell anemia, two DNA fragments are observed because the DNA has the site where MstII cuts. In a patient with sickle cell anemia the enzyme does not cut the DNA so only one DNA fragment is observed.
Today Southern blotting has been largely replaced by real-time PCR to answer the same experimental questions. Real-time PCR detects and quantifies a gene or DNA sequence of interest by recording DNA abundance throughout the amplification process, rather than just at the end as in standard PCR. This allows the researcher to compare different DNA sequences to see which is more abundant in a sample. Whereas Southern blotting is labor intensive and requires a large amount of high-quality DNA, real-time PCR has several advantages including easier automation, higher throughput screening, and a lower requirement for the amount of DNA used saving the researcher both time and resources (3). Still, researchers generating transgenic animals may choose Southern blotting to screen founder transgenic animals for the inserted transgene because Southern blots have a lower false positive rate than real-time PCR (4).
Restriction mapping and modern sequencing
 Restriction mapping informs on DNA sequence through the use of restriction enzymes that cut DNA at specific sites (5). Imagine that you have a genome with unknown sequence. In the era before sequencing was commonplace, researchers used restriction mapping to investigate DNA sequence content by comparing the fragments created by different restriction enzyme digests. Incubating a DNA sample of unknown sequence with restriction enzymes and then running the products on an agarose gel allows a researcher to properly orient specific fragments of a DNA sequence of interest to create a physical map of the DNA. Starting in the 1970s, restriction mapping was commonly used to map DNA and examine genomes (6). The knowledge gained through these experiments allowed researchers to identify the location of genes. With this knowledge, researchers were able to target specific genes for deletion allowing for the study of their function. This approach to studying DNA sequences helped researchers at a time when sequencing was still in the early days of development. Today, researchers use both Sanger and next-generation sequencing techniques to answer similar experimental questions (see below).
Restriction mapping informs on DNA sequence through the use of restriction enzymes that cut DNA at specific sites (5). Imagine that you have a genome with unknown sequence. In the era before sequencing was commonplace, researchers used restriction mapping to investigate DNA sequence content by comparing the fragments created by different restriction enzyme digests. Incubating a DNA sample of unknown sequence with restriction enzymes and then running the products on an agarose gel allows a researcher to properly orient specific fragments of a DNA sequence of interest to create a physical map of the DNA. Starting in the 1970s, restriction mapping was commonly used to map DNA and examine genomes (6). The knowledge gained through these experiments allowed researchers to identify the location of genes. With this knowledge, researchers were able to target specific genes for deletion allowing for the study of their function. This approach to studying DNA sequences helped researchers at a time when sequencing was still in the early days of development. Today, researchers use both Sanger and next-generation sequencing techniques to answer similar experimental questions (see below).
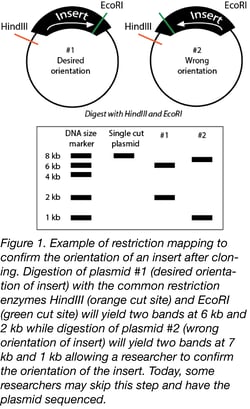
Importantly, although their role has changed, restriction enzymes are certainly not dead yet! Labs still keep restriction enzymes on hand for cloning and analytical digests. A researcher can test the orientation of an insert in only a few hours at low cost with restriction mapping (Figure 1). Though some labs may prefer to send DNA directly for sequencing (more on that below), restriction mapping may still be used to screen multiple colonies before selecting a smaller subset to sequence.
Sequencing gels and next-generation sequencing techniques
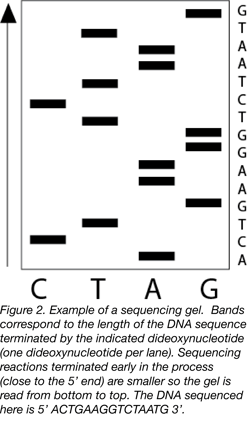 Introduced in 1977, Sanger sequencing, also known as chain-terminating or dideoxy sequencing, was a major breakthrough for determining the order of nucleotides in a string of DNA (7). Sanger sequencing uses modified nucleotides, called dideoxynucleotides (ddNTPs), lacking the 3’ hydroxyl group required for the extension of a DNA strand (8). Adding a small amount of a single radiolabelled ddNTP to a synthesis reaction along with all four regular nucleotides results in the synthesis of DNA strands of all possible lengths because extension is terminated upon the incorporation of a ddNTP. Researchers perform four different reactions, one for each radiolabelled ddNTP, and then run these reactions out in separate lanes of a gel. Since the sequencing gel will separate the DNA based on size, the resulting gel should ultimately contain radioactive bands that correspond to every nucleotide in your DNA sequence, based on which ddNTP was added and when. Compiling the identity and location of a given nucleotide, a researcher (or a simple computer program) can then reproduce the DNA sequence (Figure 2).
Introduced in 1977, Sanger sequencing, also known as chain-terminating or dideoxy sequencing, was a major breakthrough for determining the order of nucleotides in a string of DNA (7). Sanger sequencing uses modified nucleotides, called dideoxynucleotides (ddNTPs), lacking the 3’ hydroxyl group required for the extension of a DNA strand (8). Adding a small amount of a single radiolabelled ddNTP to a synthesis reaction along with all four regular nucleotides results in the synthesis of DNA strands of all possible lengths because extension is terminated upon the incorporation of a ddNTP. Researchers perform four different reactions, one for each radiolabelled ddNTP, and then run these reactions out in separate lanes of a gel. Since the sequencing gel will separate the DNA based on size, the resulting gel should ultimately contain radioactive bands that correspond to every nucleotide in your DNA sequence, based on which ddNTP was added and when. Compiling the identity and location of a given nucleotide, a researcher (or a simple computer program) can then reproduce the DNA sequence (Figure 2).
Accuracy and accessibility led to the widespread use of Sanger sequencing. Building upon this technology, radiolabelled nucleotides were replaced with fluorescently labeled ones allowing for fluorometric-based detection. This increased sequencing speed ultimately leading to what are generally considered first generation DNA sequencing machines (9). Today, similar technologies, sometimes referred to as sequence DNA as it synthesized by electronically recording fluorescence activity as fluorescently labeled nucleotides are incorporated, a method called pyrosequencing.
The drive for faster, higher throughput and less expensive sequencing fuels a sequencing revolution that still prevails. Sequencing whole genomes or profiling transcriptomes (RNAseq) require sequencing of many samples at once for later comparison. These next-generation sequencing techniques do not typically rely on Sanger sequencing but instead use methods like pyrosequencing that are parallelized to sequence thousands of reads at the same time. Other applications of sequencing, such as confirming the results of a cloning experiment, require only one or a few sequencing reactions and may use Sanger sequencing.
Looking to the future
Southern blotting, restriction mapping and sequencing gels are classic techniques that allowed for many seminal scientific discoveries (5, 8). While newer techniques have largely replaced these, these techniques still have useful applications and have informed many current techniques. For example, restriction mapping led to recombinant DNA technologies that are now commonplace in laboratories.
As scientific technologies rapidly progress, some common lab techniques we use now may soon become obsolete. For instance, the popular genome-engineering tool, CRISPR, may one day make techniques used to generate transgenic mouse lines, such as BAC transgenic or traditional knockins/knockouts, a thing of the past. In the sequencing world, one up and coming innovation is nanopore-based DNA sequencing which reads the nucleotides of a DNA strand as it slips through a tiny pore by measuring differences in ion flow caused by the different electrochemical properties of nucleotides (8). Compared to older technologies nanopore-based DNA sequencing machines are relatively small meaning that DNA sequencing could move from sequencing cores to individual labs and even out into the field. Current versions are already commercially available although they are reported to have high error rates (10). It is projected that nanopore-based sequencing technologies could one day sequence an entire genome for under $1,000 (10). The future of sequencing should bring efficient, low cost methods for sequencing large amounts of DNA quickly and accurately.
We're sure to see many improvements in a variety of biological technologies as both time and research progress. These will bring faster, higher throughput, and potentially more accurate techniques to the hands of biologists throughout the world. Let us know your favorite "Historical Lab Technique" in the comments section below!
Many thanks to our guest blogger Krissy Lyon!
 Krissy Lyon is a PhD candidate in Neuroscience at Harvard University currently studying the serotonergic system and behavior.
Krissy Lyon is a PhD candidate in Neuroscience at Harvard University currently studying the serotonergic system and behavior.
References
1. Southern, Edwin Mellor. "Detection of specific sequences among DNA fragments separated by gel electrophoresis." Journal of molecular biology98.3 (1975): 503-517. PubMed PMID: 1195397.
2. Chang, Judy C., and Yuet Wai Kan. "A sensitive new prenatal test for sickle-cell anemia." New England Journal of Medicine 307.1 (1982): 30-32. PubMed PMID: 6176866.
3. Hoebeeck, Jasmien, Frank Speleman, and Jo Vandesompele. "Real-time quantitative PCR as an alternative to Southern blot or fluorescence in situ hybridization for detection of gene copy number changes." Protocols for Nucleic Acid Analysis by Nonradioactive Probes (2007): 205-226. PubMed PMID: 17332643.
4. Haruyama, Naoto, Andrew Cho, and Ashok B. Kulkarni. "Overview: engineering transgenic constructs and mice." Current Protocols in Cell Biology (2009): 19-10. PubMed PMID: 19283728. PubMed Central PMCID: PMC2743315.
5. Roberts, Richard J. "How restriction enzymes became the workhorses of molecular biology." Proceedings of the National Academy of Sciences 102.17 (2005): 5905-5908. PubMed PMID: 15840723. PubMed Central PMCID: PMC1087929.
6. Danna, Kathleen, and Daniel Nathans. "Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae." Proceedings of the National Academy of Sciences 68.12 (1971): 2913-2917. PubMed PMID: 4332003. PubMed Central PMCID: PMC389558.
7. Sanger, Frederick, Steven Nicklen, and Alan R. Coulson. "DNA sequencing with chain-terminating inhibitors." Proceedings of the National Academy of Sciences 74.12 (1977): 5463-5467. PubMed PMID: 271968. PubMed Central PMCID: PMC431765.
8. Heather, James M., and Benjamin Chain. "The sequence of sequencers: The history of sequencing DNA." Genomics (2015). PubMed PMID: 26554401. PubMed Central PMCID: PMC4727787.
9. Hunkapiller, Tom, et al. "Large-scale and automated DNA sequence determination." Science 254.5028 (1991): 59-67. PubMed PMID: 1925562.
10. Feng, Yanxiao, et al. "Nanopore-based fourth-generation DNA sequencing technology." Genomics, proteomics & bioinformatics 13.1 (2015): 4-16. PubMed PMID: 25743089 . PubMed Central PMCID: PMC4411503.
Resources on the Addgene Blog
- Learn More about Restriction Cloning
- Get Tips on Analyzing DNA Sequencing Results
- Check out Plasmid Cloning by PCR
Resources on Addgene.org





Leave a Comment