The field of induced pluripotent stem cells (iPSCs) has been around for 10 years. In that time, scientists have used almost all available approaches for generating iPSCs. The generation of iPSCs is relatively simple in concept: ectopically express a cocktail of stem cell reprogramming factors and wait for cells to de-differentiate. However it’s difficult, especially as a newbie reprogrammer, to decide which method to use. This post provides a brief overview of reprogramming methods with the goal of helping readers choose a strategy suited to their research.
Find Plasmids for Stem Cell Research
| Delivery Method | Mechanism | Efficiency | Safety | Pros | Cons |
| MMLV-derived Retrovirus | Integrating | High | Low | Efficient and Stable |
Genome integration Insertional mutagenesis, transgene reactivation, residual expression |
| Lentivirus | Integrating | High | Low | Efficient and Stable |
Genome integration Insertional mutagenesis, residual expression |
| piggyBac | Excisible | Medium | Medium | Generates transgene-free and vector-free cells |
Genome integration Need to sequence cells to verify excision didn’t introduce mutations |
| Adenovirus | Non-integrating | Low | Medium |
Generates transgene-free and vector-free cells No genomic integration |
Inefficient |
| Sendai Virus | Non-integrating | Medium | Medium |
Generates transgene-free and vector-free cells No genomic integration |
Can be difficult to fully remove virus from cells |
| Plasmid | Non-integrating, usually | Medium | Medium |
Generates transgene-free and vector-free cells Limited genomic integration |
Multiple transfections required Need to sequence to verify there’s no genomic integration |
| Replicating EBNA1 episome | Non-integrating | Medium | Medium |
Generates transgene-free and vector-free cells No genomic integration |
Inefficient Need to verify loss of episome |
| Minicircle | Non-integrating | Medium | Medium |
Generates transgene-free and vector-free cells No genomic integration |
Inefficient Need to verify loss of minicircle |
| RNA Delivery | DNA-Free | High | High | Transgene-free and vector-free | Multiple transfections required |
| Protein Delivery | DNA-Free | Low | High | Transgene-free and vector-free. |
Slow and inefficient Difficult to purify reprogramming proteins. |
Table 1: Key Features of Different Methods for Generating iPSCs. One important note: in this table, reprogramming efficiencies ranking as low, medium or high is based on the reprogramming efficiencies reported in González et al. A percentage is not listed because many variables (cell type, criteria used, etc.) can effect reprogramming rates and such a number maybe misleading.
Integrating iPSC delivery methods
Overall, integrating viral vectors efficiently and reliably generate iPSCs, but they have several safety concerns. First, they require the use of potentially harmful viral particles that express oncogenes, such as Myc. Viral vectors also have the largest genomic footprint of iPSC generating methods due to the risk of insertional mutagenesis. Random integration also creates heterogeneous iPSC cell lines, which can complicate comparisons made between lines. Incomplete silencing of transgenes is a concern as well, and reactivation of Myc or other oncogenes after differentiation has been linked to tumor formation in iPSC-derived and iPSC-transplanted mice. Cre-deletable or Tet-inducible lentiviruses address some of these concerns, but overall integrating viral systems currently lack the safety required for translational use.
Viral Vectors
- Retroviral: Some of the first iPSCs were generated with Moloney murine leukemia virus (MMLV)-derived retroviruses. Retroviral vectors, such as pMXs or pMSCV, infect dividing cells, and have a higher efficiency of generating iPSCs compared to other delivery methods. Retrovirus has a cloning capacity of ~8 kb, so either a single polycistronic vector or multiple single-gene vectors can be used to package reprogramming factors. An important milestone of reprogramming is the silencing of reprogramming transgenes and the up-regulation of corresponding endogenous pluripotency genes. Retroviral vectors are prone to incomplete silencing of reprogramming transgenes, which leads to incomplete reprogramming. Additionally, lingering expression or re-expression of viral transgenes in iPSC-derived cells can interfere with their differentiation potential. Finally, retroviruses are prone to insertional mutagenesis and random integration creates variation between iPSC clones. Retroviruses are not considered safe for clinical applications.
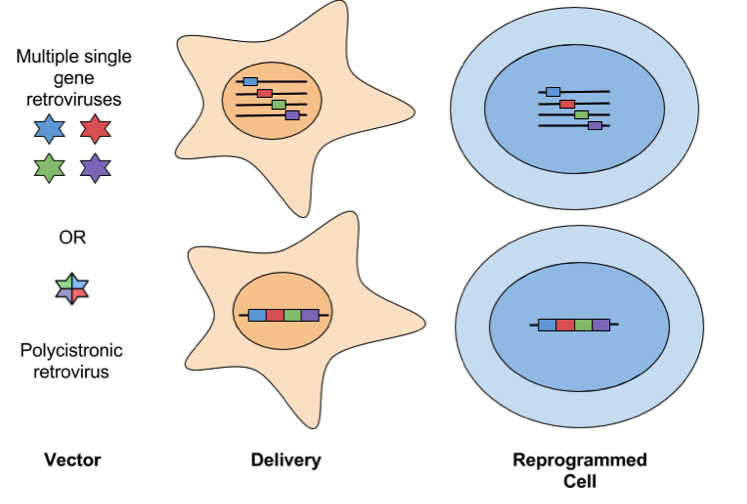
- Lentiviral: Lentiviral vectors infect dividing and nondividing cells and have a larger cloning capacity (8-10 kb) than other retroviral vectors. This makes it possible to use a polycistronic cassette that expresses multiple reprogramming factors driven by one promoter. While lentivirus has similar rates of reprogramming when compared to MMLV-derived retroviruses, it is not as well repressed following reprogramming. This can be overcome by using a Tet-inducible virus to allow for controlled expression of reprogramming factors. Check out the links at the end of the post for a list of Tet-inducible reprogramming vectors available from Addgene. Another drawback is that lentiviral vectors insert in different genetic locations in each transduced cell, so there’s unavoidable heterogeneity between clones. This can complicate comparisons made between samples, i.e. transgene expression and hence the extent of reprogramming could be affected by the location of insertion. Lentiviral insertion can also disrupt the expression of tumour suppressor genes and/or oncogenes, potentially leading to the presentation of a cancer-like phenotype in infected cells. Like retroviruses, lentiviral delivery is not considered safe for clinical applications.
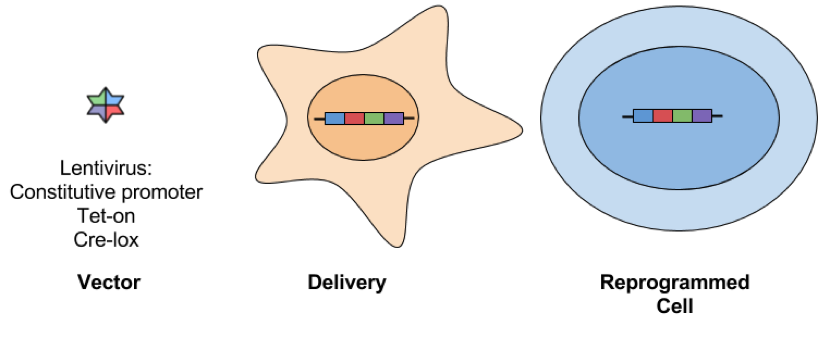
- Excisable
- Cre-lox lentiviral vectors: This approach is the same as using a standard lentiviral vector, but transient expression of Cre recombinase allows for deletion of the inserted transgenes that are flanked by loxP sites. While there is only a small loxP scar behind at the site of excision, this approach is still not considered safe for therapeutic applications.
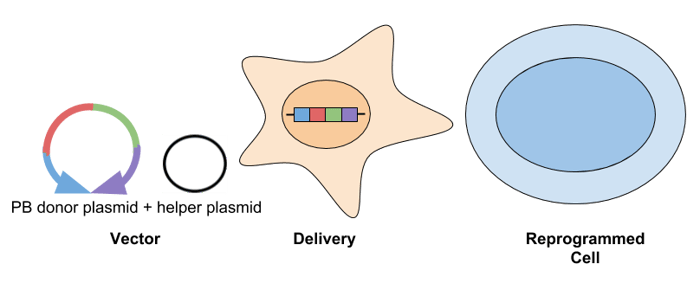
- piggyBac (PB) transposon: The PB transposon is a mobile genetic element that transposes via a “cut and paste” mechanism between a vector and chromosomal DNA. PB has a cloning capacity of ~9-14 kb. This system is transfected into cells as a donor plasmid containing the transposon, along with a helper plasmid that expresses the transposase. The donor plasmid contains the gene(s) of interest flanked by terminal repeat sequences, which the PB transposase recognizes. Following reprogramming, transgenes can be removed by re-expressing the transposase. A challenge of using PB to generate iPSCs is that genomic alterations can occur at the transposon insertion site, so sequence-verification is required.
- Cre-lox lentiviral vectors: This approach is the same as using a standard lentiviral vector, but transient expression of Cre recombinase allows for deletion of the inserted transgenes that are flanked by loxP sites. While there is only a small loxP scar behind at the site of excision, this approach is still not considered safe for therapeutic applications.
- Excisable
Non-integrating iPSC delivery methods
Viral Vectors
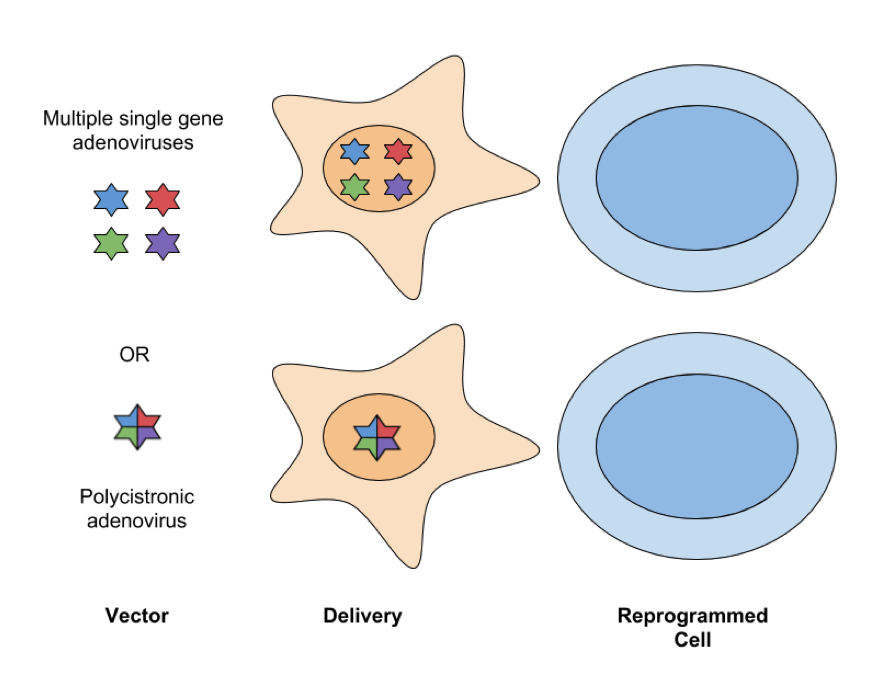
- Adenovirus: Adenoviral vectors infect dividing and nondividing cells and have an ~8kb packaging capacity. With this packaging capacity, reprogramming factors can be delivered either as a single polycistronic transgene or with four different adenoviruses, each expressing one factor. These vectors don’t integrate into the genome and are instead lost by dilution via cell division. A drawback to this approach is it has lower levels of efficiency at generating iPSCs, usually several orders of magnitude lower than retroviruses; however, because they are less likely to cause insertional mutagenesis, adenoviral vectors are considered a safer way to express reprogramming factors for therapeutic applications.
- Sendai viral vectors: Sendai virus is a single stranded, negative sense RNA virus. It’s a member of the Paramyxoviridae family of viruses, which also includes measles and mumps. Sendai transduces a wide range of cell types and replicates in the cytoplasm independent of the cell cycle. A challenge of using Sendai is that since it’s replication competent, it’s difficult to eliminate the virus from all cells, even after many passages. Ban et al developed a temperature sensitive Sendai virus which is removed by culturing cells at 38℃, but screening cells for the presence of transgenes is still an important control. Additionally, Sendai virus doesn’t rely on promoters for transcriptional regulation, so it requires a different approach for regulating transgene expression. Check out Sano et al to learn how microRNAs can be used to regulate Sendai virus transgene expression.
Transient episomal delivery
Non-integrating methods have a smaller genetic footprint compared to integrating approaches. These methods eliminate the risk of insertional mutagenesis, the presence of a genetic scar, and incomplete silencing of transgenes. Overall, non-integrating approaches are safer than integrating methods, with RNA and protein delivery considered the safest since there’s minimal risk of lingering expression of reprogramming factors.
- Non-replicating
- Plasmids: Generating iPSCs with plasmid-based expression requires serial transfection of 1 or 2 plasmids that express the reprogramming factors of interest. The advantages of this method are that it’s relatively simple to implement and doesn’t require time-consuming production of virus. In theory this is a non-integrating approach; however, in practice integration can occur. Okita et al describes a protocol for generating iPSCs by plasmid transfection where 2 of 11 clones tested had plasmid integration. Another challenge is that multiple transfection makes it difficult to control the dose of plasmid the cells receive over the whole reprogramming period and the plasmid will get diluted faster when cells are actively dividing. The typical drawbacks of using transfection will still exist: transfection efficiency is cell-type dependent and larger plasmids have lower rates of transfection.
- Minicircles: Minicircles are like mini-plasmids. They contain only a eukaryotic promoter and the cDNA(s) to be expressed and they don’t integrate or replicate. Their small size leads to higher transfection efficiencies and they tend to express for longer periods of time than traditional plasmids due to lower activation of DNA-silencing mechanisms. While minicircles are typically smaller than traditional plasmids, Jia et al generated iPSCs using a ~14.5kb minicircle expressing a 2A peptide polycistronic cassette comprised of OCT4, SOX2, LIN28, NANOG, and a GFP reporter. Minicircles are removed from cells by dilution with each cell division, but it can still take several passages for the minicircle to be completely removed.
- Plasmids: Generating iPSCs with plasmid-based expression requires serial transfection of 1 or 2 plasmids that express the reprogramming factors of interest. The advantages of this method are that it’s relatively simple to implement and doesn’t require time-consuming production of virus. In theory this is a non-integrating approach; however, in practice integration can occur. Okita et al describes a protocol for generating iPSCs by plasmid transfection where 2 of 11 clones tested had plasmid integration. Another challenge is that multiple transfection makes it difficult to control the dose of plasmid the cells receive over the whole reprogramming period and the plasmid will get diluted faster when cells are actively dividing. The typical drawbacks of using transfection will still exist: transfection efficiency is cell-type dependent and larger plasmids have lower rates of transfection.
- Replicating
- oriP/Epstein-Barr nuclear antigen-1 (EBNA1) based episomes: These plasmids carry the origin of replication (oriP) element and the cis-acting EBNA-1 from Epstein-Barr virus. EBNA-1 binds oriP and allows for replication of the plasmid in mammalian cells and assists in tethering the vector to the cell’s chromosomes. With the addition of a selectable marker, this system allows for the oriP/EBNA1 plasmid to be maintained as a stable extrachromosomal piece of DNA. Reprogramming efficiency is lower when compared to other methods. This could be a reflection of the large size of the plasmid or silencing of the plasmid by DNA methylation. Another downside of using EBNA1 based vectors is that the plasmid can hang around for awhile, even after selection is removed. Yu et al found that ⅔ of subclones generated from EBNA1 iPSCs still had episome DNA present.
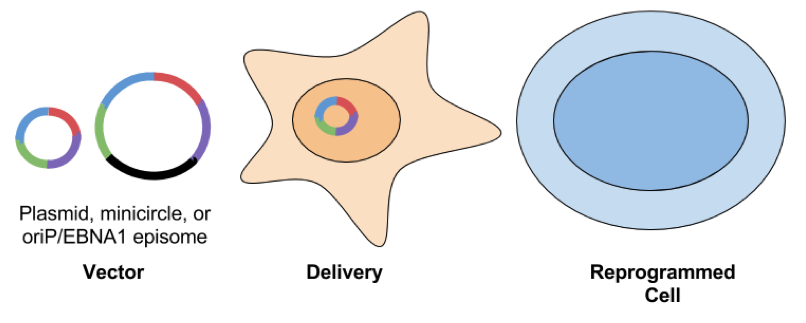
- oriP/Epstein-Barr nuclear antigen-1 (EBNA1) based episomes: These plasmids carry the origin of replication (oriP) element and the cis-acting EBNA-1 from Epstein-Barr virus. EBNA-1 binds oriP and allows for replication of the plasmid in mammalian cells and assists in tethering the vector to the cell’s chromosomes. With the addition of a selectable marker, this system allows for the oriP/EBNA1 plasmid to be maintained as a stable extrachromosomal piece of DNA. Reprogramming efficiency is lower when compared to other methods. This could be a reflection of the large size of the plasmid or silencing of the plasmid by DNA methylation. Another downside of using EBNA1 based vectors is that the plasmid can hang around for awhile, even after selection is removed. Yu et al found that ⅔ of subclones generated from EBNA1 iPSCs still had episome DNA present.
DNA-Free Methods
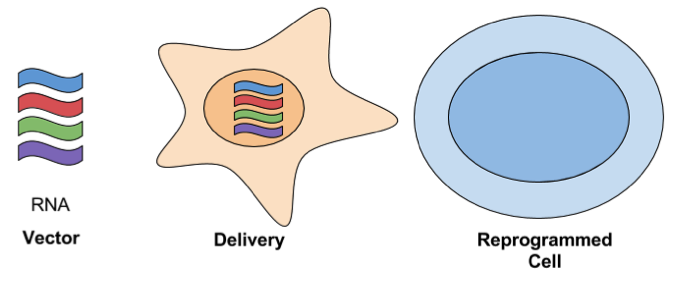
- RNA Delivery: For this approach, in vitro transcribed RNAs are serially transfected using a cationic vehicle. The following synthetic modifications are used to protect the RNAs from cellular defenses against ssRNA: phosphatase treatment, substituting cytidine with 5-methylcytidine, and/or substituting uridine with pseudouridine. The key advantage of using RNA to reprogram cells is that it’s simple and efficient. It has a high efficiency of reprogramming when compared to other methods, even when compared to integrating viral approaches. It’s also has high marks for safety.
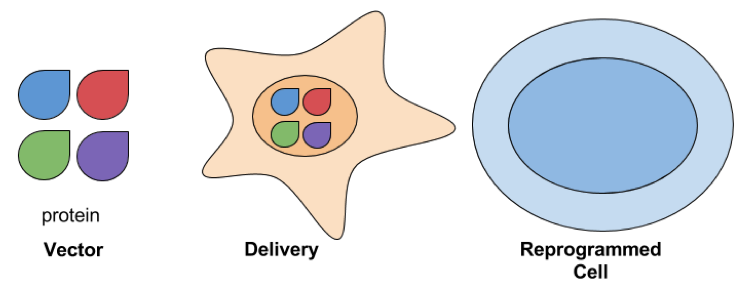
- Protein Delivery: Delivering reprogramming factors as protein is another way to avoid exposing cells to exogenous genetic material. Proteins are delivered to cells by fusing them with peptides that help mediate their transduction, such as the polyarginine peptide described in Zhou et al and Kim et al. This is considered a safer approach for developing therapeutic applications, but it can take a long time to reprogram cells. In Kim et al, cells were exposed to reprogramming proteins for 8 hours a week for 6 weeks, and still required further culturing to generate iPSC colonies. Reprogramming efficiencies are also low and it’s difficult to purify the reprogramming proteins needed to perform these experiments.
Assays for iPSCs Characterization
Once generating iPSC clones, it’s important to determine how stem cell-like they behave. iPSCs are typically tested with molecular-based assays first. These methods less rigorously assesses pluripotency, but they also take less time to complete. Morphology is a simple and fast to evaluate. iPSCs should form compact colonies that are comprised of cells with a large nucleus, large nucleoli, and little cytoplasm. Positive staining for alkaline phosphatase, expression of embryonic proteins, and promoter demethylation and expression of endogenous pluripotency genes (i.e. Oct4) are other measures of reprogramming. If a retrovirus was used, then silencing of this vector is another mark of successful reprogramming.
If cells contain molecular hallmarks of reprogramming, then they are typically assessed with more rigorous functional tests. Cells can be differentiated in vitro to form embryoid bodies which are compact balls of loosely organized tissues that express markers of all three germ layers (ectoderm, mesoderm, endoderm). Measuring pluripotency of mouse cells typically involves making chimeras by injecting iPSCs into blastocysts and implanting them into female mice. Generating chimeric offspring shows the ability of the iPSC to contribute to all three germ layers, and the ability of theses chimaeras to produce all-iPSC derived offspring demonstrates the ability of the iPSCs to form functional germ cells. The highest stringency test for mouse iPSCs, tetraploid complementation, involves injecting tetraploid blastocysts with iPSCs and measuring their ability to direct the development of a mouse.
Since many of these functional tests aren’t possible with human iPSCs, the the ability to generate teratomas is the gold standard instead. Teratoma formation requires injecting iPSCs into immunodeficient mice and looking for tumors that contains cells from all three germ layers.
Choosing a reprogramming method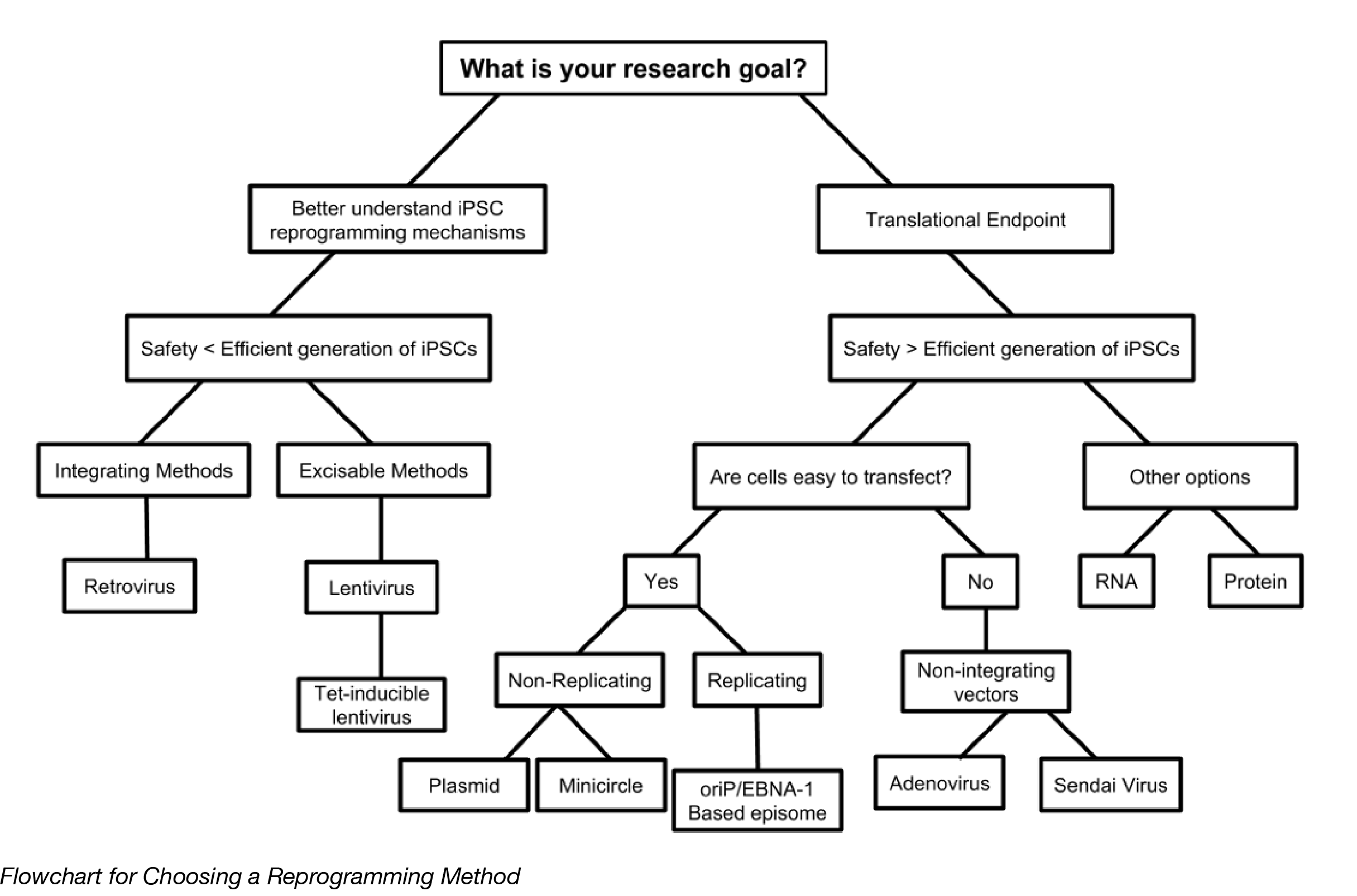
Most iPSC research falls into two categories: 1) studies focused on better understanding the mechanisms of reprogramming, and 2) studies with clinical endpoints. In the first scenario, robust and efficient generation of iPSCs is the number one priority, with safety taking lower priority. Genomic integration is less of a concern, so viral vectors are better suited for these situations. The second scenario requires higher levels of safety and as little chance of genomic alteration as possible, with efficiency being the trade-off. Non-integrating approaches, such as episomes, RNA delivery, and protein delivery are better suited for these types of studies. What cells are reprogrammed will also affect the outcome, since not all cell types are easily attainable, or easy to reprogram. Basically, there’s no universal method that can handle all applications of iPSCs, but there should be at least one method that can help you with your research goals.
Are you a new or experienced reprogrammer? Which methods do you have experience using? Let us know about your iPSCs experiences in the comments below!
References
1. Sano, Masayuki, Minoru Iijima, Manami Ohtaka, and Mahito Nakanishi. "Novel Strategy to Control Transgene Expression Mediated by a Sendai Virus-Based Vector Using a Nonstructural C Protein and Endogenous MicroRNAs." Plos One 11, no. 10 (2016). PubMed PMID: 27764162. PubMed Central PMCID: 5072705.
2. Sommer, Cesar A., and Gustavo Mostoslavsky. "The evolving field of induced pluripotency: Recent progress and future challenges." Journal of Cellular Physiology 228, no. 2 (2012): 267-75. PubMed PMID: 22767332.
3. Kim, Dohoon, Chun-Hyung Kim, Jung-Il Moon, Young-Gie Chung, Mi-Yoon Chang, Baek-Soo Han, Sanghyeok Ko, Eungi Yang, Kwang Yul Cha, Robert Lanza, and Kwang-Soo Kim. "Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins." Cell Stem Cell 4, no. 6 (2009): 472-76. PubMed PMID:19481515. PubMed Central PMC: 2705327
4. Hockemeyer, Dirk, and Rudolf Jaenisch. "Induced Pluripotent Stem Cells Meet Genome Editing." Cell Stem Cell 18, no. 5 (2016): 573-86. PubMed PMID: 27152442. PubMed Central PMCID: 4871596.
5. González, Federico, Stéphanie Boué, and Juan Carlos Izpisúa Belmonte. "Methods for making induced pluripotent stem cells: reprogramming à la carte." Nature Reviews Genetics 12, no. 4 (2011): 231-42. PubMed PMID: 21339765.
Methods figures were adapted from this reference.
1. Malik, Nasir, and Mahendra S. Rao. "A Review of the Methods for Human iPSC Derivation." Methods in Molecular Biology Pluripotent Stem Cells, 2013, 23-33. PubMed PMID: 23546745. PubMed Central PMCID: 4176696.
2. Yusa, Kosuke, Roland Rad, Junji Takeda, and Allan Bradley. "Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon." Nature Methods 6, no. 5 (2009): 363-69. PubMed PMID: 19337237. PubMed Central PMCID: 2677165.
3. Jia, Fangjun, Kitchener D. Wilson, Ning Sun, Deepak M. Gupta, Mei Huang, Zongjin Li, Nicholas J. Panetta, Zhi Ying Chen, Robert C. Robbins, Mark A. Kay, Michael T. Longaker, and Joseph C. Wu. "A nonviral minicircle vector for deriving human iPS cells." Nature Methods 7, no. 3 (2010): 197-99. PubMed PMID: 20139967. PubMed Central PMCID: 2892897.
4. Stadtfeld, M., and K. Hochedlinger. "Induced pluripotency: history, mechanisms, and applications." Genes & Development 24, no. 20 (2010): 2239-263. PubMed PMID: 20952534. PubMed Central PMCID: 2956203
5. Robinton, Daisy A., and George Q. Daley. "The promise of induced pluripotent stem cells in research and therapy." Nature 481, no. 7381 (2012): 295-305. PubMed PMID: 22258608. PubMed Central PMCID: 3652331
6. Shao, L., & Wu, W. (2010). Gene-delivery systems for iPS cell generation. Expert opinion on biological therapy, 10 2, 231-42. PubMed PMID: 20088717. PubMed Central PMCID: 2811526
Additional Resources on the Addgene Blog
- Learn about Methods to Reprogram Peripheral Blood Cells
- Check Out Our Interview with Stem Cell Researcher Darrell Kotton
- Four Factors that Differentiate the Stem Cell Field
Additional Resources on Addgene.org
- Visit our Stem Cell Guide Page
- Find reprogramming plasmids here
- Check out the Yamanaka paper that started the field of iPSCs
- Find gene-specific plasmids (including reporters and RNAi) for:
- Check out these plasmids if you’re interested in Tet-inducible lentiviral expression of OSKM reprogramming factors:
| Mouse Expression Vectors | Human Expression Vectors |
| Carey et al | Hockemeyer et al |
| Brambrink et al | Maherali et al |
| Stadtfelt et al |
Topics: Stem Cells, Other





Leave a Comment