The natural CRISPR locus of a bacteria host encodes multiple guide RNAs (gRNAs) on a single array to target the genome of the invading phage pathogen. Over the past decade, CRISPR tools have leveraged such host-defense mechanisms to enable multiplex gene editing in a variety of cells and organisms. However, lengthy genetic payloads and insufficient transcription on the array have limited the scalability and efficiency of multiplex gene editing. Moreover, existing multiplex strategies have been facing difficulties in pairing with base editing and prime editing approaches. Recently, Xue Sherry Gao’s lab at Rice University has developed DAP arrays for efficient multiplex base editing (MBE) and multiplex prime editing (MPE) with only minimal payloads.
DAP array
Although CRISPR-Cas12a, with abilities to process and release its own gRNAs from a single array, has been a top choice for multiplex gene knockout or regulations, it lacks proper functions to enable efficient base editing and has yet to be developed for prime editing, both and their derived tools are leading the edge of the precision gene editing. CRISPR-Cas9, the key component to enable efficient base editing and prime editing, is unable to process its own array but is the key component to enable efficient base editing and prime editing. How do you then set up an MBE system that contains a double-win for both efficiency and multiplexity, with these constraints?
The Gao lab first thought to enable MBE by using Cas12a fused base editors (dCas12a-BE). However, the poor editing efficiencies they found led them back towards Cas9 fused base editors (nCas9-BE), as Cas9 is mechanistically more favored for efficient base editing. They then worked to develop a strategy that would independently generate multiple gRNAs for nCas9-BE, or any other kinds of CRISPR tools, without compromising their strategic advantages in gene editing, while keeping the system to minimal size.
To do so, they harnessed the tRNA processing mechanism, which is conserved in all living organisms and has previously been used for multiplex Cas9-nuclease editing. Typically, a single array encoding the tandemly assembled tRNA-gRNAs is placed downstream of a lengthy promoter, which is used to drive the expression of the array. After expression, the individual gRNAs are released by endogenous tRNA processing machineries, specifically, RNaseZ and RNaseP. Although the Gao Lab demonstrated efficient MBE with such a strategy, the need for lengthy promoters, such as U6 or EF1α promoters, has limited the number of gRNAs that can be packaged into viral vectors for potential in vivo applications or multiplex CRISPR screening.
Using tRNAs
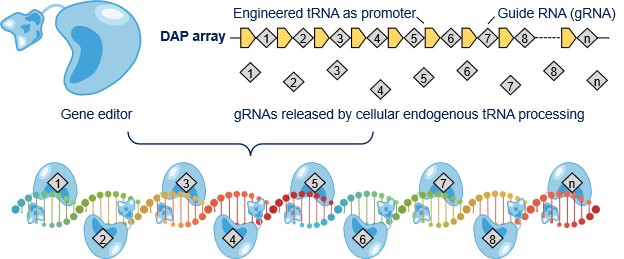
Luckily, a fun feature of tRNA was fully unleashed by the Gao Lab’s work. Led by PhD student Qichen Yuan, the Gao Lab demonstrated that the tRNA-gRNA array can drive the expression of itself, without using any additional promoter, which is followed by tRNA processing to release individual gRNAs for gene editing. They named this new array architecture the DAP array, short for “drive-and-process.”
To optimize DAP use, they developed a generalizable method to acquire the most effective DAP array with a given tRNA (derived from human or plant source), by tuning the length of 5’ leader sequences of a given tRNA to achieve the highest efficiencies of multiplex editing at each site. A 75-nt human cysteine tRNA stood out from their initial testing. It outperformed other tRNA candidates (either from human or plant sources) in both the strength as a promoter and the activity to release gRNAs from DAP array.7
Using DAP arrays with a variety of base editors, the Gao Lab successfully demonstrated up to 31-loci MBE with over 50% averaging editing efficiency in human cells. MBE with DAP array also exhibited reduced Cas9-dependent off-target editing and do not cause higher Cas9-independent off-target editing in comparison with single-site editing. Encouraged by the results of their MBE efforts, the Gao Lab also engineered a DAP array for prime editing, achieving up to 3-loci MPE with similar high efficiencies, which is scalable for a higher number of MPE.
Therapeutic potential
To explore the therapeutic potential of the DAP array, the Gao Lab packaged MBE and MPE elements for deliveries via adeno-associated virus (AAV) and lentivirus, demonstrating simultaneous editing of genes against multiple genetic diseases, such as muscular dystrophies, heart disease, and type 2 diabetes, across four different types of human cells, including T cells and hematopoietic stem and progenitor cells. Now with the DAP array, it is possible to use the state-of-art base editor, prime editor, or any CRISPR-based gene editors, for correcting mutations or installing functional genes to study and potentially treat polygenic diseases. This could also enable more convenient multiplex genome engineering of crops or allow for sophisticated CRISPR screenings that would perturb the unprecedented number of genes with a single viral vector.
 |
|
Fig. 1: Schematic of multiplex gene editing with DAP array |
Use
DAP arrays can enable efficient multiplex gene editing, such as base editing, prime editing, and nuclease editing, in virtually any cell type or organisms of interest with only minimal genetic payloads. Different from existing multiplex array architectures, including previous tRNA-gRNA arrays, that rely on an upstream lengthy promoter to express the gRNA array, DAP arrays use tRNA itself, as short as 75-nt, to both drive and process the tRNA-gRNA arrays, which is scalable and efficient in plasmid and viral vectors. In applications when repetitive tRNA sequences must be avoided, different tRNAs with optimized 5’ leader sequences can be used together in DAP array. To construct DAP array for multiplex gene editing applications, we provide detailed manual and online design examples of using Addgene plasmids hCtRNA_FT (#186715) and hCtRNA_VT (#186716).
 |
|
Fig. 2: Comparison of previous representative dual-gRNA architectures with DAP array |
Qichen Yuan is a PhD student in the lab of Xue Sherry Gao at Rice University.
References and Resources
More resources on the Addgene blog
CRISPR 101: Multiplex expression of gRNAs
Four Base Editing Reporters to Monitor and Enrich Editing in Real Time
Prime Editing: Adding Precision and Flexibility to CRISPR Editing
References
Yuan Q, Gao X (2022) Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat Commun
Topics: CRISPR







Leave a Comment