This post was contributed by guest bloggers Albert Cheng and Mark Wanner.
CRISPR-Cas9 offers a leap forward for genome editing, providing researchers with greatly enhanced accuracy, efficiency, and versatility. It has led to a tremendous acceleration of biomedical research, allowing for the modeling of human disease mutations in experimental model systems with previously unthinkable speed and precision. Furthermore, the ability to excise detrimental mutations and introduce functional sequences—as is being investigated with dystrophin/Duchenne muscular dystrophy at this time—is potentially transformative for human clinical care for some Mendelian diseases. 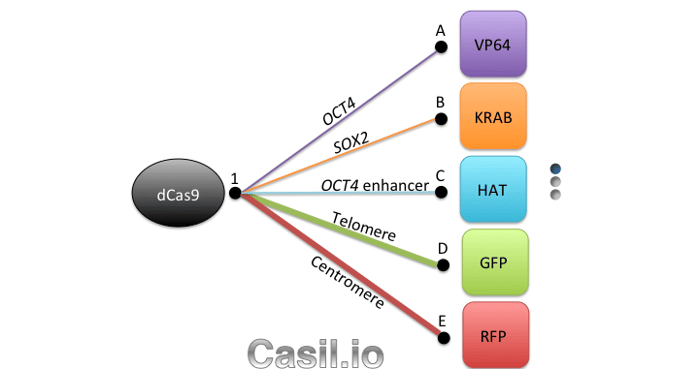
The early applications of CRISPR/Cas9
Nonetheless, as powerful as CRISPR-Cas9 has been in the lab and as promising as it may be for clinical applications, it was initially limited to a couple of functions. It was like an early smart phone with only one word processing app—revolutionary and the first of its kind but lacking the ability to do anything other than cut/delete and copy/paste in the genome. The Cas9 nuclease has since been altered to a nuclease-dead Cas9 (dCas9) by introducing mutations in its nuclease domains. dCas9 no longer cleaves DNA but still binds to specified genomic sequences with a guide RNA, functioning as a programmable DNA binding protein. dCas9 can be fused to different effector proteins to mediate functions at genomic loci, like a smartphone that can now run several more apps. The expanded functions of this “CRISPR 2.0” include gene regulation, epigenetic editing, DNA tagging for imaging, and more. The downside of CRISPR 2.0? It can only perform one function at a time because, in most cases, effector proteins are directly fused to dCas9.
CRISPR-Cas9 combinatorial functionality enabled by Casilio
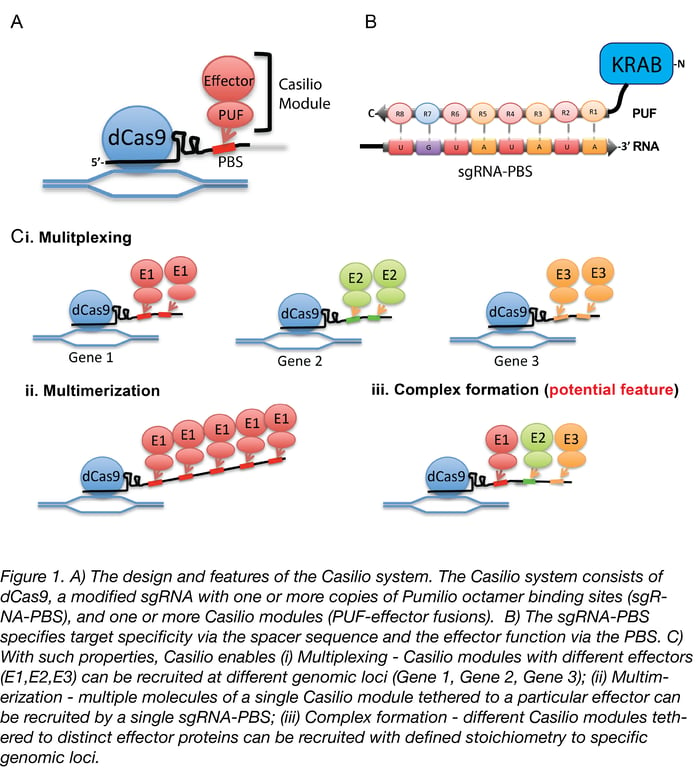
Although CRISPR 2.0 has proven quite useful for research, genes don’t work in isolation, and investigating gene networks and complex diseases that involve dysfunction across multiple genes requires a higher level of functionality. Which brings us to what we consider CRISPR 3.0: Casilio. Casilio is a powerful platform that allows CRISPR/Cas9 to multitask and easily adopt new functions, like a cutting-edge smart phone that can run multiple apps at a time and can install new apps from a constantly growing app collection. It is a hybrid construction of CRISPR-dCas9 and effectors fused to the programmable Pumilio (PUF) RNA-binding domain. PUF domains contain eight sequence repeats, each programmable by altering two specific amino acid di-residues to recognize one of the four RNA bases. With Casilio, dCas9 acts as a clamp that keeps a gRNA that has been modified to bind your desired combination of PUF-effector modules (a.k.a. “Casilio modules”) localized to your genomic region of interest (see Figure 1). Because you can recruit different PUF-effector modules via virtually any of the 48 Pumilio sequences, Casilio achieves multiple functions using a single dCas9 and an expandable library of PUF-effector modules.
The components required for using Casilio are dCas9 (pAC1445 or existing dCas9 expression vectors available through addgene.org); at least one sgRNA-PBS (single-guide RNAs appended to one or more copies of Pumilio binding sites); and Casilio modules (fusion proteins consisting of effector domain(s) fused to PUF domains programmed to recognize a specific 8-nt Pumilio binding site). To help others start exploring the near-limitless possibilities for Casilio, we offer many modules through Addgene, including modules for gene activation, gene repression, chromosome tagging (fluorescence), histone acetyltransferase modules, cloning vectors for PUF fusions, and more (see table 1).
Table 1: Casilio Modules at Addgene
| Gene Activation | Gene Repression | Epigenetic Editing | Chromosome Tagging |
| pAC1355 | pAC1451 | pAC1417 | pAC1446 |
| pAC1356 | pAC1449 | pAC1418 | pAC1448 |
| pAC1357 | pAC1415 | pAC1447 | |
| pAC1358 | pAC1416 | pAC1402 | |
| pAC1393 | pAC1450 | pAC1403 | |
| pAC1392 | pAC1404 | ||
| pAC1411 |
Example applications of Casilio
Using Casilio it is possible to recruit effector proteins to a modified single guide RNA (sgRNA-PBS) with different numbers of octamer Pumilio binding sites (PBS). As we show in our open-access paper, Casilio can multiplex PUF-tethered effectors, allowing for precise, simultaneous genome manipulations at multiple loci. In the paper, we showed simultaneous activation of OCT4 and repression of SOX2. We also demonstrated two-color labeling of telomeres and centromeres.
Another powerful potential application is the assembly of stoichiometrically defined protein complexes. This enables researchers to study complexes involved in gene regulation. By generating Casilio modules and using sgRNAs appended to different numbers and types of Pumilio binding sites, it will be possible to investigate the effects of inclusion or exclusion of specified components of a complex. It will also be possible to study the importance of stoichiometry for complex function—e.g., recruiting x molecules of component A versus y molecules of component B—and the roles that stoichiometry can play in epigenetic modifications at a target site.
Future additions to the Casilio system
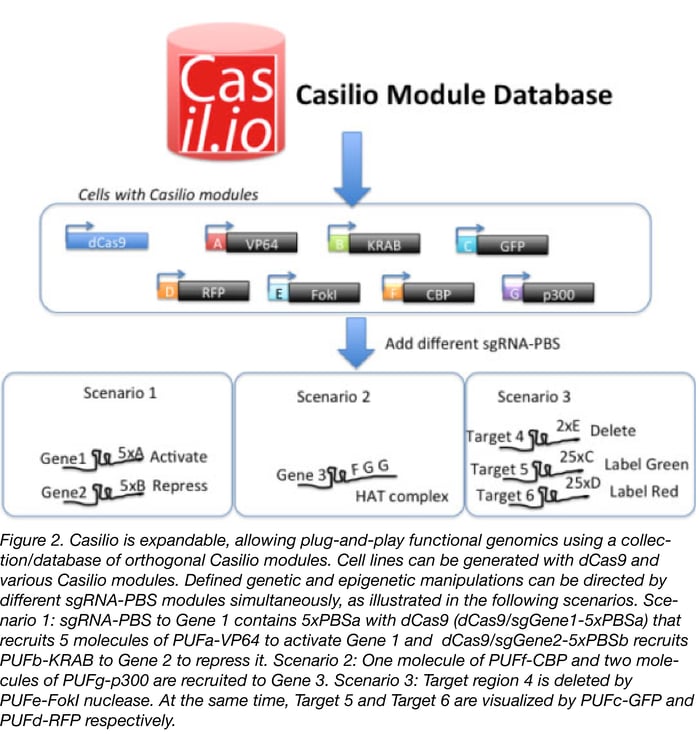
Although our current Casilio expression vectors are for transient transfection, we provide gateway donor plasmids containing the ORFs for the Casilio modules. These can be shuttled into any gateway compatible destination vectors appropriate for your cell or model systems. We are also developing a lentiviral expression vector collection of Casilio modules (lentiCasilio) to make it easy to deliver Casilio to your cell types of choice using lentivirus. LentiCasilio will be available at Addgene soon. We will also expand the collection and a database of orthogonal Casilio modules, each binding a unique PUF code. Future modules will be orthogonal, allowing plug-and-play combination of Casilio modules for functional genomics (see Figure 2).
We hope that Casilio will aid researchers in a variety of different fields and invite “developers” to build upon the Casilio platform. With this concept of a CRISPR-operating system (CRISPR-OS), we hope to see a future where “there’s a Casilio for that” for any genomic research experiment.
For updates and more details about Casilio, please visit http://casil.io and the Addgene Casilio page.
Many thanks to our guest bloggers!

Albert Cheng, Ph.D., is an assistant professor at The Jackson
Laboratory for Genomic Medicine, with a research focus on engineering
better, more powerful ways to investigate the genome, epigenome, and
transcriptome. You can follow him on Twitter @albertwcheng.
 Mark Wanner (@markgenome on Twitter) is the associate director for
Mark Wanner (@markgenome on Twitter) is the associate director for
research communications at The Jackson Laboratory and writes about all
aspects of genetics and genomics and their impact on clinical
progress.
References
1. Cheng, Albert W., et al. "Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling." Cell research (2016). PubMed PMID: 26768771. PubMed Central PMCID: PMC4746610.
Additional Resources on the Addgene Blog
- Brush up on Your CRISPR basics with Our CRISPR 101 Posts
- Read about Other Applications of CRISPR
- Learn How to Design Your gRNA
Additional Resources on Addgene.org
- Read Our CRISPR Guide
- Find Other Plasmids from the Cheng Lab
- Deposit Plasmids from Your Lab
Topics: CRISPR, Other CRISPR Tools






Leave a Comment