We are excited about our new partnership with Allele Biotechnology which allows researchers to deposit plasmids containing the fluorescent protein mNeonGreen. This fluorescent protein joins mTFP1 and mWasabi, as fluorophores from Allele Biotechnology that now can be deposited at Addgene. What makes these fluorescent proteins unique and what can they be used for? Let’s take a look!
mNeonGreen: a yellow-green fluorescent protein
mNeonGreen is a yellow-green fluorescent protein that was derived from the tetrameric yellow fluorescent protein (LanYFP) from the marine invertebrate Branchiostoma lanceolatum in 2012. Using structural modeling and structure-guided direct evolution, Nathan Shaner and colleagues from The Scintillon Institute, Allele Biotechnology, Florida State University, Indiana University, and the Karolinska Institute developed mNeonGreen via 21 substitutions to LanYFP and the addition of the enhanced GFP (EGFP)-type termini.
mNeonGreen was reported as the brightest monomeric green or yellow fluorescent protein at the time. It is 1.5 to 3 times brighter than the most commonly used GFPs and YFPs. Its excitation maxima is at 506 nm and its emission maxima is at 517 nm. Because mNeonGreen wavelengths lie between the green and yellow fluorescent protein wavelengths, it can be imaged either with standard green fluorescent protein filters or yellow fluorescent protein filters, with no or minimal reduction in emission readout, respectively.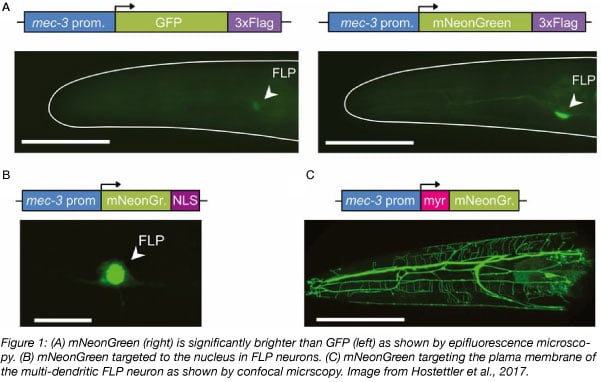
mNeonGreen can be used as a fusion tag for a wide range of applications from traditional imaging to FRET and stochastic single-molecule super resolution imaging. mNeonGreen also has little sequence identity to other common fluorescent proteins and therefore can be used in immunoprecipitation experiments with other fluorescent proteins.
Find mNeonGreen plasmids here!
mTFP1: a monomeric teal fluorescent protein
mTPF1 is a monomeric version of the tetrameric cyan fluorescent protein from the coral Clavularia sp. protein CFP484. Hui-wang Ai and colleagues from the University of Alberta and University of Oregon created mTFP1 by transforming E. coli with a gene library of approximately 484 variants, including some potential tetramer-breaking mutations. The library was then screened through successive rounds of mutagenesis for the brightest teal fluorescent proteins that were photostable and had a high 480/530 nm emission ratio. They arrived at a protein which had a total of 31 amino acid replacements relative to CFP484. Because the excitation and emission maxima were red-shifted (462 nm and 492 nm, respectively), they gave this a teal classification rather than cyan and named the protein mTFP1.
The researchers recommend using mTFP1 with a 445/30 nm excitation filter, a 470 nm beamsplitter, and a 495/30 nm emission filter. This combination results in a 2.6-fold increase in signal relative to mCerulean imaged with a standard CFP set. mTFP1 can be used as a FRET donor to mCitrine and mOrange or for live cell imaging of fusions and FRET reporters. mTFP1 is also insensitive to physiologically relevant changes in pH.
mWasabi: a bright green fluorescent protein
The team that developed mTFP1 further sought to identify why this protein was blue-shifted compared to its parent protein CFP484. Collaborating with scientists from Florida State University, they used a mutagenesis approach and found that specific amino acid changes favored blue-shifting more than others. During this process, they identified mWasabi, a bright, monomeric green fluorescent protein.
In vitro, mWasabi is 1.6-fold brighter than EGFP and is photostable. It also has narrow excitation and emission peaks (max excitation at 493 nm and max emission at 509 nm), making it useful in multicolor labeling particularly with fluorophores that excite in the violet range. For example, mWasabi can be used with Sapphire (excitation maximum 399 nm) because it does not significantly excite at 400 nm while EGFP does.
One of the most notable things about these proteins is that they are monomeric fluorescent proteins. Fluorescent proteins that oligomerize often affect the localization and/or function of the protein that they are fused to. The researchers constructed 22 different mTFP1 fusions to both the C- and N- termini of the fluorescent protein and found that the fusion protein localized to their expected locations. An analysis of mWasabi gave similar results.
We hope this blog post helped you see why certain fluorescent proteins are best suited for certain studies. If you have plasmids containing mNeonGreen, mTFP1, or mWasabi that you would like to share with the scientific community, please deposit the plasmids at Addgene!
References
Ai, Hui-wang, et al. "Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging." Biochemical Journal 400.3 (2006): 531-540. PubMed PMID: 16859491. PubMed Central PMCID: PMC1698604.
Ai, Hui-wang, et al. "Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging." BMC biology 6.1 (2008): 13. PubMed PMID: 18325109. PubMed Central PMCID: PMC2292683.
Shaner, Nathan C., et al. "A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum." Nature methods 10.5 (2013): 407. PubMed PMID: 23524392. PubMed Central PMCID: PMC3811051.
Additional resources on the Addgene blog
- Read our fluorescent protein blog posts
- Find blog posts about choosing fluorescent proteins for your research
- Subscribe to the Addgene blog
Resources on Addgene.org
- Learn about the Michael Davidson fluorescent protein collection
- Check out Addgene’s fluorescent protein plasmids and resources
- Find fluorescent protein kits
Topics: Fluorescent Proteins, Fluorescent Imaging





Leave a Comment