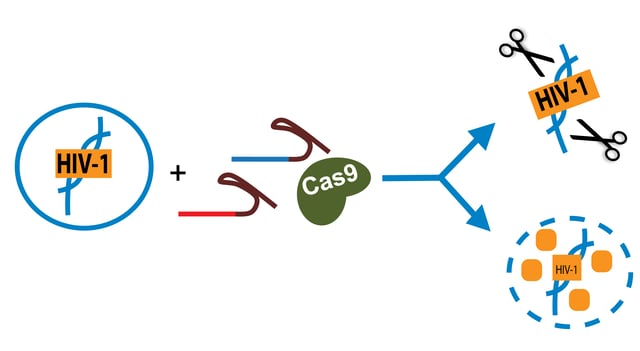
 Over 25 million people worldwide are currently infected with the lentivirus HIV-1. Today, HIV-1 can be controlled with antiviral therapies such that the virus is undetectable in the blood. But the virus doesn’t completely disappear; it just hides in latently infected cells. To truly cure HIV-1, researchers need to vanquish these hidden viral reservoirs, and CRISPR may be the way to accomplish this tough job! Kamel Khalili’s lab at Temple University has demonstrated two potential strategies for CRISPR-HIV therapeutics - one using dCas9-SAM to activate HIV-1 transcription and destroy infected cells, the other using wild-type Cas9 to remove the HIV-1 genome from infected cells. Read on to learn how CRISPR can take on HIV-1 in vitro, and what obstacles must be overcome for clinical success.
Over 25 million people worldwide are currently infected with the lentivirus HIV-1. Today, HIV-1 can be controlled with antiviral therapies such that the virus is undetectable in the blood. But the virus doesn’t completely disappear; it just hides in latently infected cells. To truly cure HIV-1, researchers need to vanquish these hidden viral reservoirs, and CRISPR may be the way to accomplish this tough job! Kamel Khalili’s lab at Temple University has demonstrated two potential strategies for CRISPR-HIV therapeutics - one using dCas9-SAM to activate HIV-1 transcription and destroy infected cells, the other using wild-type Cas9 to remove the HIV-1 genome from infected cells. Read on to learn how CRISPR can take on HIV-1 in vitro, and what obstacles must be overcome for clinical success.
ART and HIV-1 reservoirs
HIV-1 infects cells in the immune system, notably CD4+ T-cells, and eventually leads to acquired immunodeficiency syndrome (AIDS) in untreated individuals. Symptoms of AIDS include rapid weight loss and an increased risk of infection, including both common infections and opportunistic infections not usually seen in healthy individuals. Antiretroviral therapy (ART) can virtually eliminate plasma HIV-1, improving life expectancy and quality for HIV-1 patients. However, ART is not an HIV-1 cure. In patients who stop ART, viral levels will soon skyrocket back to pre-treatment levels due to the viral reservoirs located in latently infected cells. Although ART-treated patients lack plasma HIV-1, they are at increased risk for other chronic diseases, including dementia, gut disorders, nerve damage, and cardiac disease. This increased disease risk is attributed to latent HIV reservoirs, chronic inflammation, and negative metabolic effects of ART.
Why can’t the body mount an immune response to destroy these reservoirs? Essentially, the immune system doesn’t see any threat to which it should respond. Latently infected cells evade immune detection because they produce very little or no viral protein. To solve this problem, two strategies have been proposed.
The first, “shock and kill”, aims to reactivate the latent HIV so that infected cells will produce viral proteins and die, either through cytotoxicity or the immune response. The second strategy is the simple removal of the HIV-1 genome from infected cells, an idea that became much more realistic with the advent of CRISPR.
Using CRISPR/Cas9 SAM to "shock and kill"
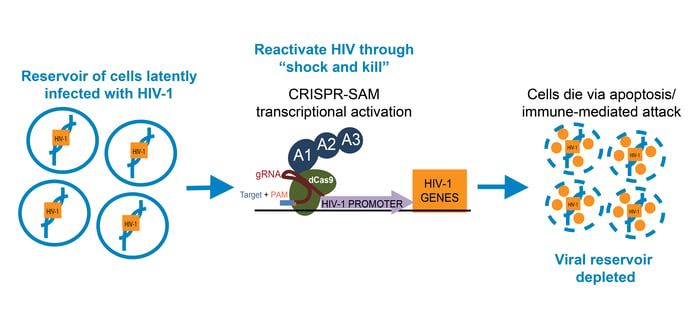
To “shock and kill” the HIV-infected reservoir, researchers have previously used histone deacetylase (HDAC) inhibitors to increase transcription of the integrated HIV-1 genome. For a targeted approach, Zhang et al. turned to the CRISPR/Cas9 Synergistic Activation Mediators (SAM) system available from Addgene. This system uses the dCas9-VP64 fusion commonly used to activate transcription, but with two additional activation domains (MS2 and p65) to enhance transcriptional activation.
Zhang et al. designed multiple gRNAs to target the 5’ long terminal repeat (LTR) of the HIV-1 genome, which acts as a promoter, and found that SAM targeting of the enhancer region near the NF-KB binding sites increased activation of HIV promoter-luciferase constructs. In multiple HIV-1 latent T cell lines, as well as a latent microglial line, treatment with CRISPR/Cas9 SAM increased the percentage of cells with activated HIV-1, measured via HIV-1 promoter-driven GFP expression. In cell lines that can produce HIV-1 toxic proteins, CRISPR/Cas9 SAM caused apoptosis, indicating a true “shock and kill” response.
Cutting HIV-1 out of the genome with CRISPR/Cas9
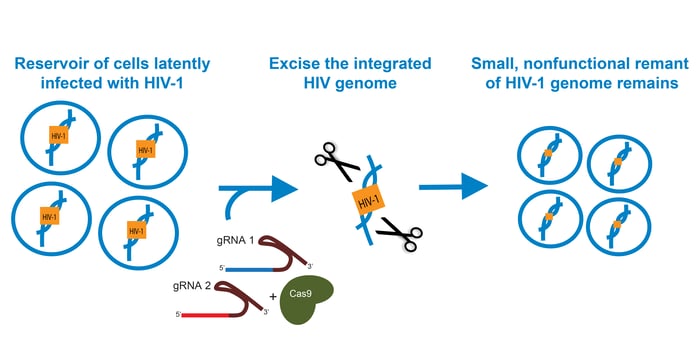
Before the introduction of CRISPR, HIV-1 genome editing with ZFNs and TALENs focused primarily on disrupting CCR5, the receptor that HIV-1 uses to enter a cell. ZFNs and CRISPR/Cas9 have also been used to create mutations in genes essential to HIV-1 replication. However, this approach could permit some viral protein production, so researchers are also interested in precisely excising the HIV-1 genome from infected cells. ZFNs were used for proof-of-concept studies, but researchers believe that using CRISPR/Cas9 could improve efficiency and reduce the probability of off-target effects.
In such a therapy, CRISPR/Cas9 would need to 1. excise the HIV-1 genome from every infected cell and 2. prevent reinfection of those cells. Kaminski et al. designed gRNAs to target the HIV-1 5’ and 3’ LTRs and expressed them along with Cas9 in the T-cell line 2D10. PCR amplification and Sanger sequencing of pooled samples showed that most of the HIV-1 genome had been excised, leaving only a small portion of the LTRs joined together. Clonal Cas9/gRNA-expressing cell populations were also immune to HIV-1 reinfection.
Importantly, no off-target effects were detected. Kaminski et al. observed no negative effects on cell viability, cell cycle progression, or apoptosis with Cas9/gRNA expression. A pooled analysis found no evidence of Cas9 cleavage at predicted off-target sites with up to 7 mismatches to the gRNA target sequence.
Kaminski et al. next tested their procedure in HIV-1 infected T-cells to see if they could be rescued by CRISPR. CD4+ T-cells were isolated from healthy individuals, expanded, and infected with HIV-1. For two HIV-1 strains, Cas9/gRNA lentiviral expression significantly reduced the HIV-1 copy number, although the efficiency differed from 48-100% between strains. In a similar experiment, CRISPR reduced the HIV-1 copy number by over 50% in isolated CD4+ T-cells from two HIV-1 infected patients. Although this is an exciting finding, it’s important to note that these HIV-1 patients were ART-naive, so this experiment models the active HIV-1 infection period rather than the ART-induced viral control found in most HIV-1 patients.
Obstacles for CRISPR HIV-1 therapy
Both of these methods represent exciting advances in preclinical HIV-1 research that should be pursued in animal models, and it’s difficult to tell if one approach might be more successful. “Shock and kill” has the advantage of killing cells infected with HIV-1 to deplete the viral reservoir, and this approach doesn’t require a functional Cas9 nuclease, so there’s no potential for aberrant DNA cleavage. Direct HIV-1 cleavage allows T-cells to survive, but preventing re-infection would require sustained expression of Cas9/gRNAs, which could result in higher rates of off-target cleavage.
In both cases, translating the in vitro work into an animal model comes with two key hurdles. The first is the delivery of the CRISPR machinery to all of the target cells. This goal may prove especially difficult given that the viral reservoir spans multiple organ systems. Both approaches may prove more successful early in the course of infection, when HIV-1 is limited to a subset of T-cells, but it is unclear if enough T-cells could be reached to ablate a well-established viral reservoir.
A second challenge for CRISPR HIV-1 therapies is sequence specificity. Whereas antiviral therapies target HIV-1 at the level of protein structure, CRISPR gRNAs require DNA sequence-specific binding. Patients’ HIV-1 genomes will need to be sequenced to determine the optimal gRNAs for either “shock and kill” or viral excision approaches, and each of these gRNAs will need to be validated for high on-target binding and low off-target binding. It’s also possible that HIV-1 may evolve resistance to CRISPR therapies through PAM or seed sequence mutations. Multiple gRNAs could be used to combat this problem, just as ART includes multiple drugs to lower the odds of developing resistance.
Shortly after Kaminski et al. published their results, Wang et al. showed that HIV-1 could escape from CRISPR/Cas9-induced modification targeting either the LTRs or essential genes. Many of these escape mutations were located near the Cas9 cut site, leading Wang et al. to conclude that some Cas9-derived indels may not ablate viral function, but rather promote resistance to CRISPR/Cas9. While these results are discouraging, it’s important to note that these experiments were done in cell culture, and the cells infected with HIV-1 only after stable expression of Cas9/gRNA. Viral production is higher in this model than in latent HIV-1 infection, which would make it easier for an “escape” virus to propagate and infect adjacent cells. It will be important to determine if CRISPR/Cas9 escape occurs in an animal model. If these escape mutations are frequent, the dCas9-mediated “shock and kill” strategy may be a better option than direct cleavage.
Despite the potential difficulties in translating these findings into a therapy, these papers present tantalizing evidence that an HIV-1 cure may be within our reach. Similar studies have shown that CRISPR can be used to combat other viral infections, notably a mouse model of Hepatitis B, a disease that infects over 250 million people worldwide. As we’ve previously seen in the CRISPR field, the lessons learned in pursuing one technological application can benefit many others and accelerate the pace of research. We at Addgene hope to see the problem of in vivo CRISPR delivery become more tractable in the next few years, as it would open up many new therapeutic possibilities for some of the world’s most common diseases.
References
Zhang Y, Yin C, Zhang T, Li F, Yang W, Kaminski R, Fagan PR, Putatunda R, Young WB, Khalili K, Hu W. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep. 2015 Nov 5;5:16277. PubMed PMID: 26538064. PubMed Central PMCID: PMC4633726
- Find SAM plasmids used in this publication at Addgene.
Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci Rep. 2016 Mar 4;6:22555. PubMed PMID: 26939770. PubMed Central PMCID: PMC4778041.
- Find CRISPR and lentiviral packaging plasmids used in this publication at Addgene.
Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Rep. 2016 Apr 5. PubMed PMID: 27068471.
Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015 Jun 2;5:10833. PubMed PMID: 26035283. PubMed Central PMCID: PMC4649911.
Khalili K, Kaminski R, Gordon J, Cosentino L, Hu W. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J Neurovirol. 2015 Jun;21(3):310-21. PubMed PMID: 25716921.
Additional Resources on the Addgene Blog
Additional Resources onf Addgene.org
- Check out Addgene’s newly updated CRISPR Guide!
- Find CRISPR Plasmids for Your Research
- Browse gRNA Design Tools
Topics: CRISPR, CRISPR Therapeutic Applications





Leave a Comment