This post was contributed by Rory Macneil, founder of Research Space.
There are many types of electronic lab notebooks (ELNs), each with its own pros and cons. All ELNs have the virtue of liberating data from paper into an electronic environment and hence making it searchable and shareable, but some ELNs provide better support for reproducibility than others.
Unfortunately, most ELNs are designed as closed ecosystems and act as data silos because they limit connectivity with other tools and resources. Thus, it’s difficult to get data out of the ELN. This means that reproducibility is limited in two ways – only data inside the ELN is reproducible, and only the highly restricted number of people who can access the ELN have the ability to attempt to reproduce the research that produced the data.
In this post I’d like to delve deeper and look specifically at the characteristics and capabilities an ELN needs to facilitate true reproducibility. The following are critical: (1) connectivity to other data sources used in research; (2) connectivity to other tools used in research; and (3) connectivity to open source data repositories. To illustrate this, let’s take a look at the RSpace ELN.
The connected approach in action: how the RSpace ELN supports reproducibility
RSpace was designed in response to requirements that emerged from intensive engagement with a wide range of people at the University of Wisconsin in 2011/12 – researchers and PIs, science librarians and data curators, IT managers, the commercialization office, the CIO’s office, and research computing. The two guiding principles emerging from the engagement with the Wisconsin research community were to: (1) prevent vendor lock in by making it easy to get data out of the ELN; and (2) facilitate inter-operability with other research tools and infrastructure in use at research universities not only today, but also in the future. Reproducibility was both a fundamental driver of these principles and has emerged as a byproduct of connectivity.
Between 2012 and 2017 RSpace was developed with these principles in mind, and in response to further input provided by the research communities at the University of Edinburgh, the University of Goettingen, and several other large research institutions. The following graphic depicts the ways in which the two guiding principles have been implemented by providing support for connectivity to external data sources, tools used in research, and data repositories. Below I describe how each of these kinds of connectivity facilitates reproducibility.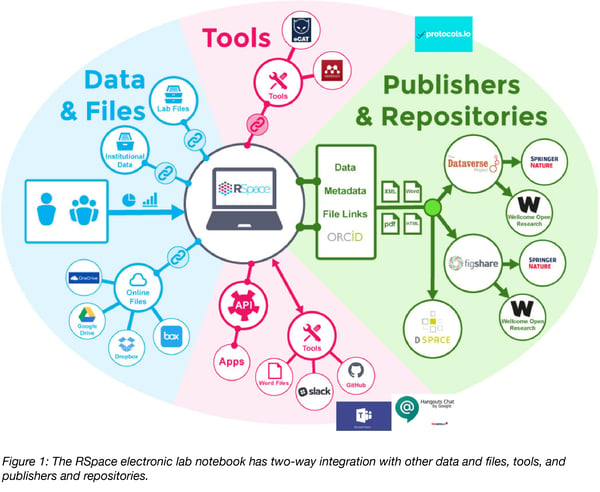
Connectivity to other data sources
The data entered into an ELN is only ever a small fraction of all data directly relevant to a particular experiment, series of experiments, or project. ‘Small’ data produced by the research group in spreadsheets, Word docs, and the like is likely stored in online file storage tools like Dropbox, Box, One Drive, and G Drive. ‘Big’ data like image banks and sequencing files is likely stored in institutional and lab servers specifically designed to hold large amounts of data. Although it may be useful to selectively bring particular items into an ELN – e.g. an image that will be re-used frequently in the group’s research – it makes no sense and is not practical to move all this data in bulk into a lightweight tool like an ELN. It is nevertheless critically important to be able to conveniently reference specific pieces of data stored externally in the record of research compiled in the ELN. Without such references the record of research is incomplete, making it difficult or impossible to reproduce. As the graphic depicts, RSpace supports linking to all of the most commonly used online file storage tools, as illustrated in this video, and to data stored on institutional and lab servers.
Connectivity to other tools used in research
Just as researchers need to access and reference external data sources, they also need to continue to make use of other tools they use in research. This includes chat apps like Slack, Microsoft Teams, and Google Hangouts Chat, resources like Github and Gitlab, and science-specific tools, e.g. sample management apps, colony management software, and method repositories like protocols.io. These tools are essential to the workflows used in producing research data, and without access to them, the record of research captured in the ELN is incomplete. Thus, to support reproducibility an ELN should have integrations with a broad range of relevant generic and science-specific tools. In addition, it should provide a modern and fully supported API that enables development of custom integrations with other tools and resources. As depicted in the graphic, RSpace is integrated with most of the tools noted above, and has a modern API. Integrations with additional tools are added on a regular basis, so the ecosystem of related tools is always growing.
Beyond the ability to continue to use other tools in conjunction with the ELN, in some cases the integrations provide a direct boost to reproducibility. For example, it’s possible to import Word documents into RSpace, maintaining the original formatting. In this way data is moved from a static and isolated vehicle and becomes part of the dynamic and connected record of research. In RSpace, document versions are maintained when the document is edited, and it’s possible to link between various documents.
A second example is the integration between RSpace and Slack, illustrated in this video. This is a two-way integration. It’s possible to send documents from RSpace to a selected Slack channel for discussion and review. It’s also possible to capture Slack conversations and import them into RSpace. The kind of informal conversations about research that can take place in Slack normally never become part of the permanent research record, yet they are crucial to understanding research and the thinking and workflows that went into it. The ability to capture the conversations and associate them with a particular experiment or project thus significantly enhances the ability of others to subsequently reproduce the research.
A third, powerful example is the integration with protocols.io, the protocol sharing repository. As illustrated in this video the integration enables RSpace users to search for protocols in protocols.io, and then link to a selected protocol and/or import and convert it into RSpace. The reverse flow is also supported, so it’s possible to re-export a protocol which has been edited back to protocols.io. Thus, RSpace becomes part of a workflow that enhances the sharing and reproducibility ecosystem protocols.io makes possible.
Connectivity to open source data repositories
Over the past decade or so a number of general purpose repositories have been launched and are now a main vehicle for making research data available for public access, review, and query. Gates Open Research and Wellcome Open Research both accept deposit of data associated with publications into one of five general purpose open source data repositories: Dataverse, Zenodo, Open Science Framework, Data Dryad, and Figshare. Any ELN that facilitates reproducibility has to make it easy to export data from the ELN directly into at least one and preferably more than one of these five general purpose repositories.
RSpace has been integrated with two of the five: Figshare and Dataverse, as well as the DSpace repository. These integrations utilize an interface that enables users to conveniently select and deposit individual documents or more complex datasets into the desired repository. They also support entry of required metadata, with the option of adding additional metadata in the repository after the deposit has been made. It’s possible to associate the depositor’s ORCID ID with the deposit, and a DOI is added to the deposit by the repository. The ability to deposit data that has already been structured in folders or notebooks in the ELN, without having to add at the time of deposit, makes it far more likely that deposits will be made. The Dataverse integration is described in this video and the Figshare integration in this video.
The ability to link from RSpace documents to external data sources (described above) further enhances the value the repository integrations, because when RSpace documents are deposited into the repositories, links in the documents to data in the external file storage resources remain live. This means that someone who wants to query and attempt to reproduce an experiment documented in RSpace using the RSpace datasets deposited in Figshare or Dataverse has access not only to the RSpace documents, but also to the associated data (spreadsheets, images, sequencing data, etc.) used and/or referenced in the experiment.
Since most ELNs, unlike data repositories, are designed to support confidentiality and limit permissions to view and edit to particular groups and/or particular individuals, the issue of third-party access to the ELN arises here. Notwithstanding access restrictions based on permissions, however, in some cases it would be possible to grant those wanting to understand and/or attempt to reproduce research relating to data deposited into a repository access to that part of the ELN from which the data came.
The ability to re-import data from repositories back into ELNs would also be useful, so that the data can be viewed – and conceivably additional work relating to reproducing the original research can be documented -- in the original context in which it was produced, collected, and analyzed.
While the many different ELNs out there can help with searchability, record-keeping, and data sharing, not all of them support reproducibility in the same way. When choosing an ELN for your research, be sure to consider how its features do and don’t support reproducibility as not all ELNs are created equal.
Many thanks to our guest blogger, Rory Macneil!

Rory is founder and CEO of Research Space, which provides the RSpace electronic lab notebook. Rory is based in Cambridge, Massachusetts, and feels privileged to be part of the vibrant life sciences research and commercialization community in greater Boston, while at the same time keeping one foot on the other side of the pond through interacting with colleagues in Edinburgh. You can follow Rory on Twitter @rory_macneil.
Additional resources on the Addgene blog
- Find tips on choosing an electronic lab notebook
- Watch a livestream of the Minisymposium on Reproducibility
- Read all blog posts about scientific sharing
Resources on Addgene.org
- Find protocols of molecular biology techniques
- Find plasmids at Addgene
- Make your work more accessible by depositing at Addgene





Leave a Comment