Transposons are sequences of DNA that can move around in a genome. In a laboratory setting, transposons can be used to both introduce genes into an organism’s genome (see figure) and to disrupt endogenous genes at the site of insertion. In both of these cases, transposons combine the advantages of viruses and naked DNA while eliminating some of the drawbacks. Specifically, viruses are able to infect and replicate in host cells, but they are susceptible to cells’ defense mechanisms. The use of non-viral vectors, like transposons, avoids many, though not all, of these defenses. For some applications of genome engineering - such as certain forms of gene therapy - avoiding the use of viruses is also important for social and regulatory reasons.
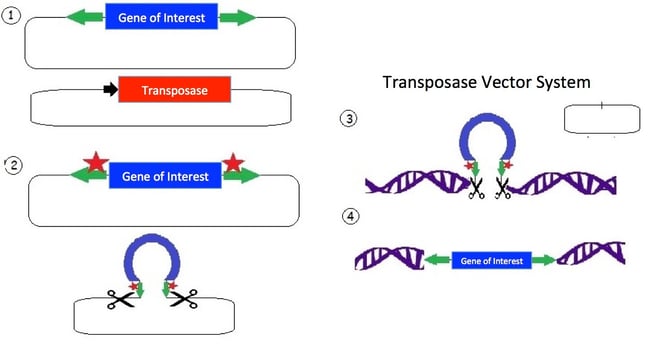
1.) The transposon consists of a mirrored set of inverted repeats (green) flanking the gene of interest (blue) in a plasmid backbone. A separate plasmid contains the transposase gene (red) for expressing the transposase enzyme.
2.) The transposase enzyme is expressed (red stars) and binds the inverted repeats (green); and an endonuclease reaction occurs which cuts the DNA.
3.) The released transposon can now bind a strand of DNA with a TA dinucleotide (there are many such sites in the human genome). The original plasmid is empty following the removal of the transposon; the plasmid is then degraded by the cell.
4.) The transposase creates a double strand break in the DNA and allows the tranposon to integrate. Additionally, the TA sequence is duplicated near the gene of interest insertion site.
The Sleeping Beauty transposon system
The Sleeping Beauty (SB) transposon system is one such system which has been designed to perform gene transfer in vertebrates. The system is comprised of 2 components: 1) an SB transposase, the enzyme required for catalysis of transposition; and 2) a transposon containing the gene-expression cassette that can translocate within the genome. The Sleeping Beauty transposase gains its name from the fact that it was recreated from a defective transposase found in fish. This transposase has been discovered in the genomes of many different fish species in a variety of non-functional forms and is believed to be millions of years old. The first functional SB system was synthesized by fusing and modifying various extinct transposases. Improvements have been made since then by increasing the consensus between a greater number of extinct transposon sequences and then testing various combinations of changes.
The basic functional version of the Sleeping Beauty transposon (SB10), developed in 1997, continues to be altered in order to improve its gene insertion efficiency in primary cells. In 2009, Zsuzsanna Izsvak’s lab screened many SB tranposase variants for hyperactivity in mammalian cells. Their efforts resulted in a new and improved SB transposase, SB100X, that is approximately 100-fold more efficient than the first-generation transposase (as determined by transposition assays of antibiotic-resistance genes conducted in HeLa cells). In their Nature Genetics paper, the lab showed that efficiencies of stable gene transfer using SB100x were comparable to stable transduction efficiencies of integrating viral vectors.
New Sleeping Beauty transposase vectors
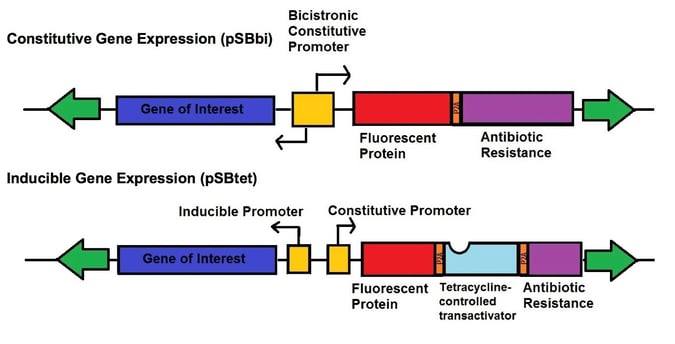
Even more recently, Eric Kowarz’s lab developed two new types of vectors for the SB transposase system - one set of vectors for robust constitutive expression (pSBbi derivatives) and one set of vectors for inducible expression (pSBtet derivatives). The constitutive and inducible expression vectors were developed into 16 variants with different selection markers (blasticidin, neomycin, puromycin, hygromycin) and fluorescent protein reporters (GFP, BFP, RFP, no FP). The vectors each contain two SfiI restriction sites with different overhangs depending upon whether they are on the 3’ or 5’ end of the gene of interest allowing for insertion in the correct orientation. These plasmids use a novel synthetic RPBSA promoter, which is made up of a fragment of the RPL13a promoter fused to a region of the RPL41 gene, to drive the selection/reporter parts of each construct. This new tool kit allows the rapid generation of stable transgenic cell lines which can robustly express your gene-of-interest, either constitutively or regulated by doxycycline.
The Sleeping Beauty transposase is proving an exciting tool for producing specific genetic mutations and gene disruptions without the use of viral vectors. In 2009, it was named Molecule of the Year. Additionally, in 2014 researchers at MD Anderson Cancer Center presented results from a clinical trial using the Sleeping Beauty transposase as a nonviral vector for CD19-directed chimeric antigen receptor (CAR) therapy. SB is a valuable tool and an alternative to CRISPRs for genome engineering whose use is only likely to grow in the coming decades.
References
1. Kowarz E, et al. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J. 2015 Feb 4. doi: 10.1002/biot.201400821. PubMed.
2. Mates L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009 Jun;41(6):753-61. Epub 2009 May 3. PubMed.
3. Singh H, et al. A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol Rev. 2014 Jan;257(1):181-90. doi: 10.1111/imr.12137. PubMed.
Find the Plasmids at Addgene
Topics: Other Plasmid Tools, Plasmids






Leave a Comment