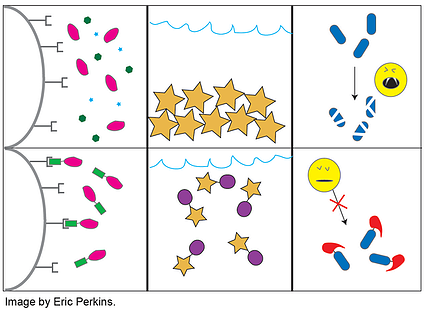
Protein tags are usually smallish peptides incorporated into a translated protein. As depicted in the accompanying cartoon, they have a multitude of uses including (but not limited to) purification, detection, solubilization, localization, or protease protection. Thus far Plasmids 101 has covered GFP and its related fluorescent proteins, which are sometimes used as tags for detection; however, those are just one (admittedly large) class of common fusion protein tags. Biochemists and molecular biologists who need to overexpress and purify proteins can face any number of technical challenges depending on their protein of interest. After several decades of trying to address these challenges, researchers have amassed a considerable molecular tool box of tags and fusion proteins to aid in the expression and purification of recombinant proteins.
Tags for stability and solubility
What are some of the hurdles to overcome in order to overexpress a recombinant protein? It is not generally in a cell’s best interest to overexpress a protein. Energy and cellular resources are being spent to make something the cell doesn’t need to make. Eukaryotes and some bacteria deploy proteosomes to degrade what the cell might consider junk protein. Though there are a number of chemical and peptide-based proteosome inhibitors, glutathione S-transferase (GST), which can be fused to recombinant proteins for one-step purification with glutathione, can also protect against proteolysis.
That’s one form of instability. Prokaryotes can also have a hard time folding eukaryotic proteins. You can get your bacteria to produce massive amounts of protein, but if it’s not folded correctly, there’s no point in crystallizing it or testing its function. Small ubiquitin-related modifier (SUMO) can help with folding and stabilization, as can maltose-binding protein (MBP). Overexpression can also lead to insolubility, and aggregated protein is not useful protein. MBP tags can help with solubility issues, but scientists may also choose to add smaller proteins, such as Thioredoxin A (TrxA) that improve disulfide bond formation in order to help keep your protein soluble.
Tags for affinity and purification
An affinity tag, generally a relatively small sequence of amino acids, is basically a molecular leash for your protein. If you’re working with an uncharacterized protein, or a protein for which a good antibody has not been developed (and just because your protein has a commercially available antibody, that doesn’t mean it’s a good one), then your first step towards detecting, immunoprecipitating, or purifying that protein may be to fuse an affinity tag to it. The FLAG, hemaglutinin antigen (HA), and c-myc tags have been the workhorses of the affinity tag world for years, and deciding on which one to use will depend on your application (see table below). The antibodies available for these tags really are good and can be used for western blots, IP, and affinity purification.
Arguably the simplest affinity tag is the polyhistidine (His) tag. Small and unlikely to affect function, His-tagged proteins can be purified using metal-affinity chromatography, usually using a Ni2+ column. Like other affinity tags, a His tag can be fused to either the N- or C-terminus of a protein. Unlike other epitope tags – which when doubled or tripled increase the tag size quickly – modifying the length a polyhistidine tract does not greatly alter the size of the tag.
Table 1: Common protein tags
| Tag | Epitope | Mass (kDa) | Function | Notes |
| CBP | KRRWKKNFIAVSAANRFKKISSSGAL | 4 | Affinity and Purification | Binding and elution steps use very moderate buffer conditions |
| FLAG | DYKDDDD or DYKDDDDK or DYKDDDK | 1 | Affinity and Purification | Good for antibody-based purification; has inherent enterokinase cleavage site |
| GST | Large Protein | 26 | Purification and Stability | Good for purification with glutathione; protects against proteolysis, but may reduce solubililty |
| HA | YPYDVPDYA or YAYDVPDYA or YDVPDYASL | 1.1 | Affinity | Frequently used for western blots, IP, co-IP, IF, flow -cytometry; can occassionally interfere with protein folding |
| HBH | HHHHHHAGKA GEGEIPAPLA GTVSKILVKE GDTVKAGQTV LVLEAMKMET EINAPTDGKV EKVLVKERDA VQGGQGLIKI GVHHHHHH | 9 | Combo | Consists of a bacterially derived in-vivo biotinylation signaling peptide (Bio), flanked by hexahistidine motifs (6xHis) |
| MBP | Large Protein | 40 | Solubility and Purification | Can improve solulibility and folding of eukaryotic proteins in prokaryotes; single step purification with amylose, but wicked huge |
| Myc | EQKLISEEDL | 1.2 | Affinity | Frequently used for western blots, IP, co-IP, IF, flow -cytometry, but rarely used for purification as elution requires low pH |
| poly His | HHHHHH | 0.8 | Affinity and Purification | Very small size, rarely affects function |
| S-tag | KETAAAKFERQHMDS | 1.8 | Solubility and Affinity | Abundance of charged and polar residues improves solubility; good for antibody-based detection |
| SUMO | ~100 amino acid protein | 12 | Stability | At N-terminus, promotes folding and structural integrity; cleavable. Not great for purification; too cleavable in eukaryotes |
| TAP | GRRIPGLINP WKRRWKKNFI AVSAANRFKK ISSSGALDYD IPTTASENLY FQGEFGLAQH DEAVDNKFNK EQQNAFYEIL HLPNLNEEQR NAFIQSLKDD PSQSANLLAE AKKLNDAQAP KVDNKFNKEQ QNAFYEILHL PNLNEEQRNA FIQSLKDDPS QSANLLAEAK KLNDAQAPKV DANHQ | 21 | Combo | See text |
| TRX | MSDKIIHLTD DSFDTDVLKA DGAILVDFWA EWCGPCKMIA PILDEIADEY QGKLTVAKLN IDQNPGTAPK YGIRGIPTLL LFKNGEVAAT KVGALSKGQL KEFLDANLAG SGSGHMHHHH HHSSGLVPRG | 12 | Solubililty | Assists in proper folding |
| V5 | GKPIPNPLLGLDST | 1.4 | Affinity and Purification | Good for antibody-based purification |
Combo and cleavage tags
Frequently, a single tag is not enough. What if you need one tag to increase solubility and one tag for purification? Or you want to combine a fluorophore with a tag that localizes your protein to the nucleus? Or you want multiple rounds of purification to get your protein as pure as possible? Vectors that offer different combinations of tags are readily available, and though adding too many tags and fusion proteins to your protein of interest would eventually get ridiculous (you generally don’t want more tag than protein), 2-3 tags is increasingly common. Tandem affinity purification (TAP) once referred specifically to a combo tag comprised of a calmodulin binding peptide (CBP), a TEV cleavage site (more on that in a moment), and 2 ProtA IgG-binding domains. TAP has since come to encompass several other tag combinations, though frequently those combinations still include at least one element from the original TAP tag. The terms dual-labeling and dual-tagging are also used. Due to their small size and the ease with which they can be added to a purification scheme, His tags are frequently combined with other tags for dual-labeling.
The problem with all these tags is that many of them serve a one-time purpose, and you don’t necessarily want them to stick around after that purpose has been served. At this point, proteases can be your friend rather your enemy. Two common tags (SUMO and FLAG) are cleaved by specific proteases without requiring the addition of an independent cleavage recognition site. In fact, SUMO cannot be used in eukaryotes because there is already too much SUMO protease around, but it is convenient when used with purified protein since the enzyme cleaves the SUMO tag in the same manner as it would have in the context of a cell. FLAG tags can be cleaved by enterokinase, which recognizes DDDDK^X, cleaving after the lysine. The efficiency of this cleavage depends on the identity of X.
A number of other proteases are available, but scientists would need to incorporate their recognition sites into their protein tag in order to use them effectively. One of the best optimized is the tobacco etch virus (TEV) protease. A TEV protease cleavage site is frequently placed between two tags being used for two rounds of purification, with the cleavage reaction taking place between column runs. The TEV protease itself, with various mutations used to increase its stability activity, can be readily purified using plasmids found in this paper (available at Addgene).
Table 2: Protease recognition sites commonly used with tags
| Protease | Recognition site | Notes |
| TEV | ENLYFQS | Cleaves between the Gln and Ser residues |
| Thrombin | LVPRGS | Cleaves between Arg and Gly residues |
| PreScission | LEVLFQGP | Cleaves between the Gln and Gly residues |
This article is not a comprehensive guide to all tags, but rather a quick overview of why scientists use tags, with a few time-tested tags and fusion proteins as examples. The tables list more common tags than are described in the post, but have been categorized to help you better assess their function. More detailed information and some protocols can be found in the references provided.
Additional resources:
- Young CL, Britton ZT, Robinson AS, Biotechnology Journal 2012 , (7): 620-634. Full Text
- A complete list of free handbooks for protein purification is provided by GE Healthcare Life Sciences.
Addgene pages and blog posts of interest:
- Which Fluorescent Protein Should I Use?
- Plasmids 101: Green Fluorescent Protein (GFP)
- Fluorescent Protein Guide
- Expression Vectors from the Berkeley QB3 MacroLab
Eric thanks his wife, Annette Sievers, Ph.D., for her tips and guidance. She is way better at protein purification than he is.
Topics: Plasmid Elements, Plasmids 101, Plasmid Tags







Leave a Comment