Addgene’s plasmids are used with a wide variety of restriction enzyme-based cloning methods. Each method has its own pluses and minuses, but Golden Gate cloning has been especially useful within both the synthetic biology and genome engineering fields. We’ll walk you through how to apply this precise and easy-to-use system to your cloning efforts.
Golden Gate cloning technology relies on Type IIS restriction enzymes, first discovered in 1996. Type IIS restriction enzymes are unique from "traditional" restriction enzymes in that they cleave outside of their recognition sequence, creating four base flanking overhangs. Since these overhangs are not part of the recognition sequence, they can be customized to direct assembly of DNA fragments. When designed correctly, the recognition sites do not appear in the final construct, allowing for precise, scarless cloning.
The cloning scheme is as follows: the gene of interest is designed with Type IIS sites (such as BsaI or BbsI), that are located on the outside of the cleavage site. As a result, these sites are eliminated by digestion/ligation and do not appear in the final construct. The destination vector contains sites with complementary overhangs that direct assembly of the final ligation product. As shown below, a fragment with 5’ overhang TGGA and 3’ overhang TCCG can be ligated into a vector containing those overhangs. Entry DNA overhangs may be present in the original plasmid (Option 1) or added using PCR-based amplification (Option 2). 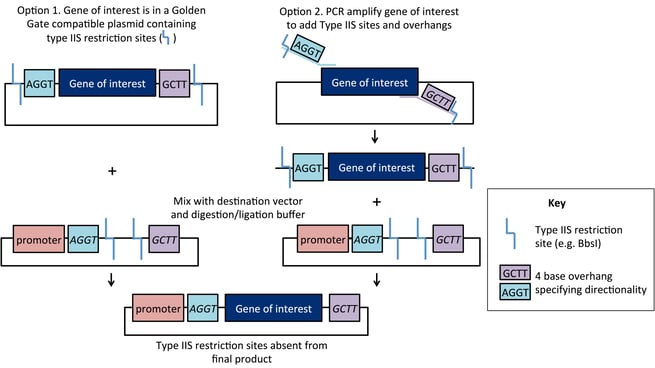
Advantages of Golden Gate cloning
Golden Gate cloning is one of the easiest cloning methods in terms of hands-on time, as digestion and ligation can be done in one 30-minute reaction. The destination vector and entry vector(s) are placed in a single tube containing the Type IIS enzyme and ligase. Although the original destination vector + insert may spontaneously religate, this transient construct retains functional Type IIS sites and will be re-digested. In contrast, formation of the desired ligation product is irreversible because this construct does not retain the enzyme recognition sites. As a result, the ligation process is close to 100% efficient. Another strength of Golden Gate cloning is its scalability. Unique 4 base overhangs can be used to assemble multiple fragments - up to 10 fragments are commonly assembled in a single reaction! These overhangs specify the desired order of fragments, and the loss of enzyme recognition sites after ligation favors formation of the construct of interest. Although efficiency may decrease with an increased number of fragments, or the ligation of very small/very large fragments, these problems can be overcome by screening a higher number of potential clones. Golden Gate assembly has a few advantages over other cloning methods. Exonuclease-based methods like Gibson assembly require 20-40 bp of homology at the ends of DNA fragments to specify assembly order, so fragments with 5’ or 3’ sequence homology cannot be assembled using this method, but can be assembled with Golden Gate. The popular Gateway cloning system produces constructs with an attB recombination scar encoding eight amino acids, but Golden Gate assembly can be designed to be scarless. Golden Gate assembly is also less expensive than many commercial cloning methods.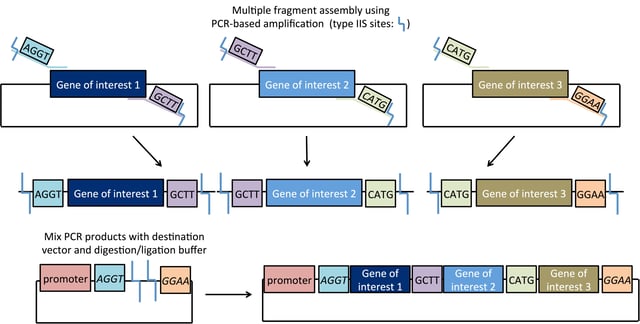
Golden Gate and synthetic biology
Synthetic biologists have leveraged the power of Golden Gate cloning into a modular cloning strategy. Sometimes referred to as MoClo, this strategy uses the Type IIS restriction enzymes BsaI and BpiI/BbsI to efficiently assemble up to six DNA fragments at a time. As with all Golden Gate-based methods, this system exploits the ability of Type IIS enzymes to cut outside their recognition site and permits DNA fragments with compatible overhangs to be efficiently assembled. Scientists can engineer unique enzyme recognition sites that flank their DNA fragment in an inverse orientation. This allows for multiple DNA components (promoters, genes, terminators, etc) to be assembled in the correct order in a single reaction. For detailed Golden Gate protocols, complete with helpful tips and tricks, see The Sainsbury Lab website or Engler & Marillonet.
Golden Gate and genome engineering
In early 2011, the Bogdanove and Voytas groups described a new Golden Gate-based technology for genome editing which allowed for the ordered assembly of multiple DNA fragments to create TAL effector nucleases. These plasmids were designed to utilize the BsaI and BsmBI Type IIS sites such that custom TAL arrays could be constructed quickly and efficiently in just a few steps. More recently, CRISPR technology has adapted Golden Gate cloning for inserting the appropriate oligonucleotides specifying a gRNA target sequence into a Cas9-containing plasmid such as pX330. This cloning strategy not only makes it easy to create a single gRNA-expressing plasmid, but it can also be adapted to express multiple gRNAs. Addgene has two Golden Gate-based gRNA assembly methods available, which allow you to efficiently clone up to 7 gRNAs into one destination vector, making multiplexing easy.
Disadvantages of Golden Gate cloning
Golden Gate cloning is not 100% sequence-independent: to avoid undesired digestion, the Type IIS site used must not be present within the fragments you seek to assemble. One way to work around this is to "domesticate" your fragment: PCR-based amplification can be used to create silent point mutations at internal recognition site(s) thus eliminating these from your gene of interest. PCR products are then digested with the Type IIS enzyme, and the mixture is ligated following a heat inactivation step. If your genes of interest or destination vector contain multiple internal restriction sites that may not be amenable to "domestication", you might want to consider using an alternative method like Gateway cloning or Gibson assembly. Another important consideration is the design of flanking overhangs. Although there are theoretically 256 distinct flanking sequences, sequences that differ by only one base may result in unintended ligation products.
Whether you are using the Golden Gate method to create CRISPR/Cas9 constructs, assemble standard plasmids parts in different combinations, or other new and exciting applications, this system is an incredibly powerful tool for cloning complicated constructs in a single, high-efficiency step.
References:
Sequential amplification of cloned DNA as tandem multimers using class-IIS restriction enzymes. Lee JH, Skowron PM, Rutkowska SM, Hong SS, Kim SC. Genet Anal.1996 Dec;13(6):139-45. PubMed.
A one pot, one step, precision cloning method with high throughput capability. Engler C, Kandzia R, Marillonnet S. PLoS One 2008;3(11):e3647. doi: 10.1371/journal.pone.0003647. Epub 2008 Nov 5. PubMed.
Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Nucleic Acids Res. 2011 Jul;39(12):e82. Epub 2011 Apr 14. PubMed.
Golden Gate cloning. Engler C, Marillonnet S. Methods Mol Biol. 2014;1116:119-31. doi: 10.1007/978-1-62703-764-8_9. PubMed.
More Resources at Addgene:
- Check out more Plasmids 101 Posts!
- Check out Addgene’s restriction cloning guide!
- CRISPR 101: Multiplex Expression of gRNAs
- Choosing a Molecular Cloning Technique
Topics: Plasmids 101, Plasmid Cloning, Plasmids







Leave a Comment