In addition to single plasmids, Addgene also distributes pooled plasmid libraries containing hundreds, thousands, or even a million plasmids. These libraries are some of Addgene’s most exciting and versatile offerings! We recently re-amplified our distribution stock of the Brunello Human gRNA library, and we thought it would be a good time to talk about the amplification and verification processes we use to ensure high-quality library distribution. You can also use these tips as a starting point when you need to amplify a library for your own experiments.
 Library amplification 101
Library amplification 101
Library amplification seems straightforward: E. coli bacteria are transformed with the library DNA, they grow and replicate, and then their DNA is harvested, much like one would prep an individual plasmid. However, lots of library DNA is needed for screening experiments, so the scale of the DNA prep is larger. And since the library contains many plasmids, care must be taken to ensure that all of these plasmids are amplified and none are lost.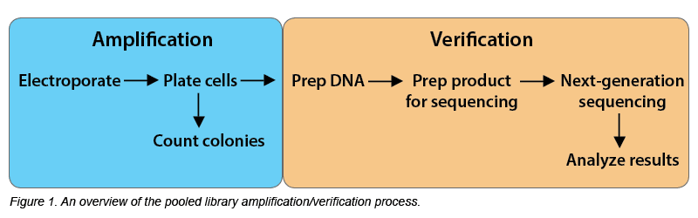
Each Addgene library has its own depositor transformation and amplification protocol, but the overall workflow is similar for most libraries. Here are the key points you’ll need to remember when undertaking library amplification and sequencing.
Maintain representation of the library during transformation
First, use enough DNA and competent bacteria to ensure all components of the library are represented. Typically, Addgene uses electrocompetent Stbl4 cells when performing amplifications, and we use electroporation to ensure a good transformation efficiency. For a typical library, Addgene usually does four to eight electroporation reactions using a total of ~400ng of library DNA. These reactions should allow us to obtain about 100 to 1,000 bacterial colonies per individual plasmid in the library.
Bacterial growth must also be optimized to maintain library diversity. If the various plasmids in the library are differently sized, smaller plasmids may be amplified preferentially, skewing the composition of the final recovered DNA. This bias can be counteracted by growing the cells on solid plates, where each transformant forms its own colony, uninfluenced by the growth of other cells. For libraries where the plasmids are all very similar sizes, like a gRNA library, this is less of a concern, but Addgene still recommends plating the transformation mixture on solid media. The colonies are then scraped off the plates and pelleted for DNA extraction.
Prepare library product for next-generation sequencing
Once the DNA has been extracted, the representation of the library should be confirmed using next-generation sequencing (NGS). For gRNA libraries, you’ll design primers to create 200-300 bp sequencing products using PCR. A diagram of the product we created for the Brunello library is shown in Figure 2. You can use a small PCR product because the plasmids in the library are identical except for the insert; there’s no need to sequence other parts of the plasmid.
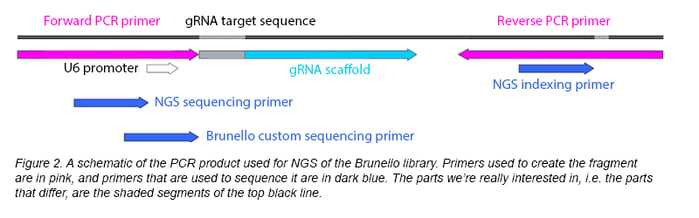
How will you know if your sequencing results represent a successful amplification? To provide a point of reference, Addgene commonly prepares NGS samples using both pre- and post-amplification DNA and generates PCR sequencing products from both. These products are then purified, quantified, and sent for sequencing.
Analyzing your sequencing results
If your sequencing is successful, your NGS provider should return results in the form of many, many short DNA sequences called reads. Typically they’ll be in a file in FASTQ format, a standard format for storing NGS reads and their corresponding sequencing quality. Generally, you want to have 100x-1000x coverage of your library: in other words, 100-1000x more reads than number of plasmids in the library. Analyzing the reads is, again, seemingly straightforward – you need to count how many times each of the library components is found in the read list. If you also sequenced the pre-amplification DNA, you’ll compare the numbers before and after. However, because there are hundreds of thousands if not millions of reads, this analysis must be done in an automated manner. Addgene commonly uses a custom script derived from the count_spacers.py file from Feng Zhang’s laboratory. There are a number of other analysis scripts available, some associated with specific libraries, but any analysis script should return the same output: a spreadsheet of read counts.
You can use these counts to confirm the representation of your library. When plotted as a histogram, the counts will give you a bell curve of the abundance of each component of your library. You should look for two things: underrepresented plasmids (plasmids that were lost or “dropped out”) and overrepresented plasmids (plasmids that were amplified preferentially).
You can also rank the plasmids by abundance, scaled to 1, to create a “cumulative wealth distribution” curve, or Lorenz curve. By calculating the area under the curve (AUC), one can gain a sense of the uniformity of the library distribution, with an AUC of 0.5 representing perfect uniformity (see Figure 3A). Addgene finds that this representation is most easily understood visually. As an example, the curves from our recent amplification of the Brunello library are also shown in Figure 3. I hope you’ll agree with me that they look very similar to each other - that’s exactly what we want!
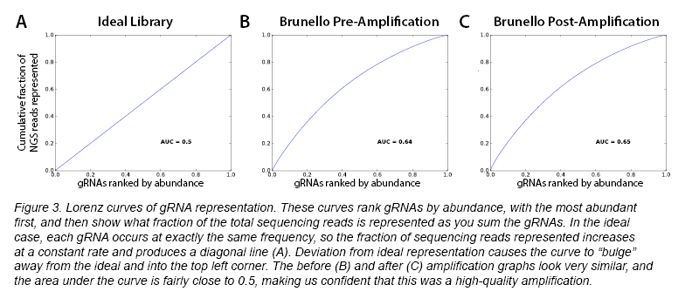 At the end of the day, it’s up to you to decide your threshold of what a good amplification looks like, but by looking at these kinds of graphs, making sure few/no plasmids have dropped out, and comparing pre- and post-amplification sequencing results, you should be able to determine the success of your amplification.
At the end of the day, it’s up to you to decide your threshold of what a good amplification looks like, but by looking at these kinds of graphs, making sure few/no plasmids have dropped out, and comparing pre- and post-amplification sequencing results, you should be able to determine the success of your amplification.
I hope this post has helped clarify some of the important considerations when using, amplifying, and verifying pooled plasmid libraries. Pooled libraries can be a powerful research tool, but their proper use requires careful amplification and maintenance of good representation. Addgene wishes you luck in your future library use and screening, and if you have any tips, input, or stories of interest about library use, let us know in the comments!
Additional Resources on the Addgene Blog
- Check out Our Plasmids 101 Topic Page
- Learn How We Use NGS Sequencing to Validate AAV Preps
- Learn More about Pooled CRISPR Libraries
Resources on Addgene.org
- Browse Pooled Libraries at Addgene
Topics: Plasmids 101, Plasmids






Leave a Comment