Hallmarks of fluorescent protein timers
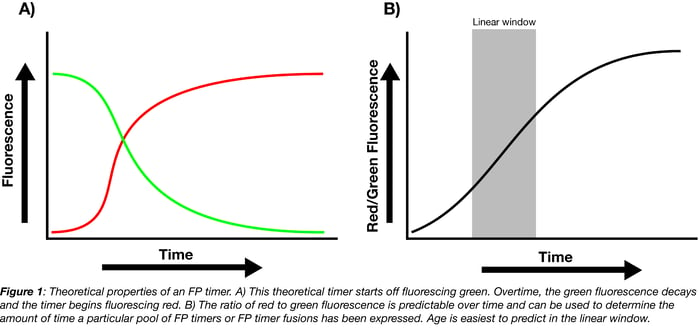
When FP timers are first expressed, they predominantly fluoresce one color but slowly mature until they predominantly fluoresce a second color. This change is usually hypothesized to be a result of chromophore oxidation. Some of the more popular timers (whether as part of protein fusions or expressed independently) originally fluoresce green or blue and, when they mature, fluoresce red. The ratio of the mature color (red) to the immature color (green or blue) within a population of timer indicates the age of the pool: the higher the ratio, the older the pool.
FP timers have been around since 2000 when Terskikh et al. reported the production of dsRed E5. This timer predictably transitions from green to red fluorescence (500 to 580 nm) over the course of 18 hours in vitro (see figure 1B in Terskikh et al.) and even displays predictable kinetics over 14 hours of expression in C. elegans embryos. However, this initial timer was a tetramer with a propensity to aggregate in cells (Tsuboi et al.), limiting its use. Nonetheless, its creators noticed a few properties that have now become hallmarks of ideal FP timers:
1. Ratiometric determination of expression time
The ratio of mature to immature fluorescence from the FP timer is dependent on the total expression time but independent of protein concentration. Calibration curves correlating this ratio to the total expression time can therefore be used to determine how long a pool of FP timers has been expressed, either in whole cells or as a pool of fusion proteins localized to a specific region of the cell. For example, if the ratio of red/green fluorescence falls within the linear window for the theoretical timer shown in Figure 1B, you can use the equation fit to this linear region to solve for total expression time.
2. Timer functionality with a single gene
Although it is possible to make a timer system composed of two separate FPs (one that matures quickly and the other slowly, see Verkhusha et al.), one benefit of the common FP timers is that you can monitor age by expressing a single FP-timer protein or protein fusion. This one protein method simplifies cloning and makes your experimental setup less cumbersome.
3. Portability across multiple systems
Terskikh et al. showed that their versatile timer could be used in vitro, in mammalian cells, in C. elegans, and in Xenopus embryos. Of course, timers may be sensitive to changes in oxygen, temperature, and pH, but timer activity can be calibrated to these different conditions, enabling the use of a given timer in many experimental settings.
Monomeric fluorescent protein timers
Since the production of the initial FP timer in 2000, researchers have realized how problematic aggregation-prone tetrameric fluorescent proteins can be. Cytotoxicity, improper localization, and decreased functionality are all possible consequences of FP aggregation, as Erik Snapp’s blog post shows. To avoid these issues, Subach et al. and Tsuboi et al. developed monomeric FP timers with less propensity to aggregate.
These monomeric FP timers (mK-GO from Tsuboi et al. and the FT series from Subach et al.) were derived from previously developed monomeric fluorescent proteins mKO and mCherry, respectively. While Tsuboi et al developed mK-GO somewhat serendipitously while attempting to enhance mKO in other ways, Subach et al made a concerted effort to develop their FT series using their knowledge of protein structure and saturation mutagenesis. mK-GO matures from green to red, and the FT series matures from blue to red (see Table 1 for emission and absorption spectra). Subach et al’s directed mutagenesis also produced the three separate timers shown in the table: Fast-FT, Medium-FT, and Slow-FT. These display progressively longer blue fluorescence maturation times and varied blue-red maturation times (Table 1). These separate proteins should be useful for monitoring cellular events occurring at varied time scales.
Table 1: Monomeric FP Timers and Associated Plasmids from Subach et al.
| Protein | Excitation (nm) | Emission (nm) | Brightness | pKa | Maturation | Plasmids |
|---|---|---|---|---|---|---|
| Fast-FT | 403 (blue), 583 (red) |
466 (blue), 606 (red) |
14.9 (blue), 6.8 (red) | 2.8/4.1 | 7.1 h (red) | |
| Medium-FT | 401 (blue), 579 (red) | 464 (blue), 600 (red) |
18.4 (blue), 5.8 (red) |
2.7/4.7 |
3.9 h (red) | |
| Slow-FT | 402 (blue), 583 (red) | 465 (blue), 604 (red) |
11.7 (blue), 4.2 (red) |
2.6/4.6 |
28 h (red) |
Applications of fluorescent protein timers
FP timers have been used to:
- Monitor dynamics of vesicle fusion and release from the plasma membrane by fusing various vesicle cargoes to FP timer mk-GO (Tsuboi et al.)
- Monitor gene expression in the developing pancreas by placing FP timer DsRed-E5 under the control of Neurog3 (a gene controlling pancreatic differentiation) in mouse embryos (Miyatsuka et al.)
- Distinguish between models for trafficking to the lysosome by fusing Medium-FT to LAMP-2A, a lysosome-associated membrane protein, and following its subcellular localization overtime (Subach et al.)
- Monitor protein expression dynamics in a synthetic circuit (Saxena et al.)
In a general sense, FP timers can be used in any situation where one wants to understand the relationship between the age of a cell, protein, or cellular structure and a particular biological event (trafficking to a subcellular location, start of gene expression, development of a cell structure, etc). FP timers should therefore find use in studies of animal development where events like the patterning of nascent tissues and the formation of limbs are correlated with changes in gene expression. In a developing embryo, for example, a researcher could determine whether or not newly synthesized or long-lived transcription factors control gross changes in gene expression as new body sections are formed. In synthetic biology, FP timers might be able to tell researchers whether old or new cells are better at producing a compound of interest, thus allowing them to optimize the compound production process.
How have you used FP timers? Are you planning on using an FP timer in a new and creative way? Let us know in the comments section below!
References
1. Terskikh, Alexey, et al. ""Fluorescent timer": protein that changes color with time." Science 290.5496 (2000): 1585-1588. PubMed PMID: 11090358.
2. Tsuboi, Takashi, et al. "Age-dependent preferential dense-core vesicle exocytosis in neuroendocrine cells revealed by newly developed monomeric fluorescent timer protein." Molecular biology of the cell 21.1 (2010): 87-94. PubMed PMID: 19889833. PubMed Central PMCID: PMC2801723.
3. Verkhusha, Vladislav V., et al. "An enhanced mutant of red fluorescent protein DsRed for double labeling and developmental timer of neural fiber bundle formation." Journal of Biological Chemistry 276.32 (2001): 29621-29624. PubMed PMID: 11408473.
4. Subach, Fedor V., et al. "Monomeric fluorescent timers that change color from blue to red report on cellular trafficking." Nature chemical biology 5.2 (2009): 118-126. PubMed PMID: 19136976.
5. Miyatsuka, Takeshi, Zhongmei Li, and Michael S. German. "Chronology of islet differentiation revealed by temporal cell labeling." Diabetes 58.8 (2009): 1863-1868. PubMed PMID: 19478145. PubMed Central PMCID: PMC2712795.
6. Saxena, Pratik, et al. "A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells." Nature communications 7 (2016). PubMed PMID: 27063289. PubMed Central PMCID: PMC4831023.
Additional Resources on the Addgene Blog
- Learn More About Fluorescent Protein Aggregation
- Catch Up On Your GFP Basics
- Get Advice on Choosing Your Fluorescent Protein
Resources at Addgene.org
- Find Fluorescent Protein Timers
- Browse All Fluorescent Protein Empty Backbones
- Check Out Our Fluorescent Protein Guide






Leave a Comment