
Protein purification can be one of the most stressful lab activities. Working with proteins requires a substantial amount of properly folded, relatively pure protein, but getting to this stage is often much easier said than done. As reviewed in our Plasmids 101 series, proteins are overexpressed from a plasmid construct, most often in special E. coli strains designed for protein expression. Cultures are then lysed and the protein of interest is purified using an affinity tag. Additional tags may be used to improve protein stability and solubility.
Determining the best way to express one’s protein of interest can save a lot of time later. Protein purification specialists recommend doing some research, notably determining the subcellular localization and the number of disulfide bonds in your protein - two parameters that affect how well E. coli can produce your protein. Structural information for many proteins is easily accessible in the RCSB Protein Data Bank and PDBsum. After acquiring this information, you can then pick an expression vector and expression system. If you find that your protein might not fold well, you should screen a few different vectors to see which gives you the most functional protein.
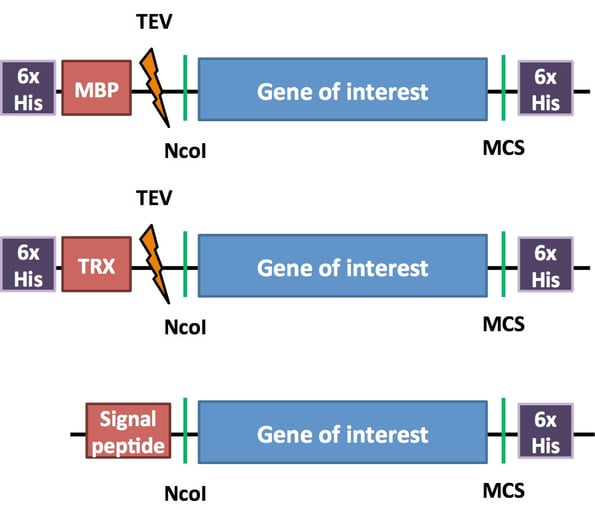
The pCri system, recently deposited to Addgene by F. Xavier Gomis-Rüth, aims to take the guesswork out of protein purification through simple and efficient multiplexing. pCri plasmids are divided into two sets, each of which includes plasmids with different varieties of tags, both N- and C-terminal. Each set of plasmids uses the same restriction enzymes for cloning (see schematic below), enabling a researcher to test multiple tags for one protein quickly and easily. These tagged plasmids also include cleavage sites should one need to remove the tag post-purification.A sample of pCri constructs: pCri-1a, -4a and -9a. 6xHis tags enable affinity purification using a Ni-NTA resin. MBP improves protein solubility and stability. TRX and signal peptide tags help proteins with disulfide bonds fold properly. If a TEV protease site is present, TEV can remove these tags from the purified protein. For more detailed information on these and other tags used in the pCri system, please see Plasmids 101: Protein Tags.
Beyond E. coli: purifying from Bacillus subtilis and Pichia pastoris
pCri also includes plasmids designed for protein expression in systems other than E. coli, specifically the bacterium Bacillus subtilis and the yeast Pichia pastoris. The improved secretory capacities of these microbes make them useful for producing difficult-to-fold proteins. In addition, P. pastoris has the capacity to glycosylate proteins and may recapitulate the post-translational modifications added to proteins in other eukaryotes better than E. coli. Plasmids designed for these systems may include a signal peptide (SP) that routes the protein through the secretory pathway to improve folding and disulfide bond formation. If one prefers expression in E. coli, the use of a thioredoxin A (TRX)-containing vector can improve disulfide bond formation.
pCri also includes a special feature for difficult-to-purify membrane proteins. pCri vector pCri-13a, designed for use in B. subtilis, contains the MISTIC (membrane-integrating sequence for translation of integral-membrane protein constructs) protein tag. MISTIC drags tagged proteins into the membrane and may improve expression of proteins that natively localize there.
pCri is both versatile and easy to use, and we hope that it will take some of the stress out of protein purification. For a more thorough introduction to pCri, please see the original publication cited below. pCri plasmids are available either individually or as a complete kit from Addgene.References
1. Goulas, Theodoros, et al. "The pCri system: a vector collection for recombinant protein expression and purification." PloS one 9.11 (2014): e112643. PubMed PMID: 25386923. PubMed Central PMCID: PMC4227841.
2. Young, Carissa L., Zachary T. Britton, and Anne S. Robinson. "Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications." Biotechnology journal 7.5 (2012): 620-634. PubMed PMID: 22442034.
Additional Resources
- RCSB Protein Data Bank - a database of detailed protein structural information
- PDBsum - visualization of the data from RSCB PDB
Topics: Plasmid Tags, Plasmids






Leave a Comment