Primary cells recapitulate the natural biology of a cell type of interest better than immortalized lines derived from the same cell type; however, their usage has been limited by technical problems. For instance, it’s much more difficult to introduce a gene of interest into primary cells, so most primary cell lines require viral infection. A new paper from Niels Geijsen’s lab suggests that primary cells may be better transduced using only protein. Read on for a description of the lab’s iTOP protein-only transduction method and its potential applications to CRISPR/Cas9 genome editing.
Protein-only transduction?
Niels Geijsen’s group in Utrecht wanted to pursue a nonviral strategy for transducing primary cells. What if, instead of supplying a DNA construct, they could simply induce cells to take up protein directly? It’s not a new idea, but it comes with a few caveats. The most common protein-only transduction strategy is to fuse a cell penetrating peptide (CPP) to a protein of interest thereby giving it the capacity to enter cells. One such CPP was discovered in the HIV protein Tat. Although CPP fusions can translocate a cell membrane, the presence of the CPP may alter protein function or localization. Thus, the usefulness of CPPs must be evaluated on a case-by-case basis.
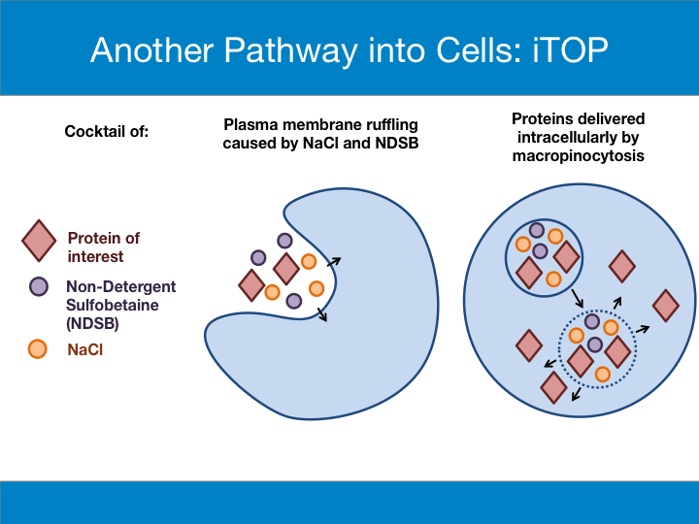
While setting up their CPP system to study protein transduction, D’Astolfo et al. made the surprising discovery that both purified CPP-tagged protein and an untagged control could enter cultured cells and activate expression of a luciferase reporter. CPP was not required for protein entry into cells! After verifying the specificity of their luciferase assay, they concluded that components in the buffer used for protein purification must be helping the untagged protein enter cells. Subsequent work found that both NaCl and non-detergent sulfobetaine 201 (NDSB-201) were required for protein delivery. D’Astolfo et al. coined the term iTOP (induced transduction by osmocytosis and propanebetaine) to describe this method of protein transduction. Later studies showed that iTOP functions through macropinocytosis.
D’Astolfo et al. tested iTOP in a series of primary cell types using Cre and various loxP reporters. In both mouse and human embryonic stem cells, they found very high recombination percentages (78-79% after two rounds of transduction.) Compared to CCP transduction, iTOP is at least four times more efficient in primary fibroblasts. iTOP also works in multiple types of primary mouse cells, including neuronal and gut stem cells, dendritic cells, embryonic fibroblasts, glia cells and neurons, and cell death post-transduction is low.
 |
| Neuronal stem cells transduced with iTOP resulting in GFP expression. Image courtesy of Diego D'Astolfo. |
CRISPR/Cas9: a potential application of iTOP
Having tested iTOP using Cre-lox recombination, D’Astolfo et al. were eager to see if iTOP would be compatible with CRISPR/Cas9 genome editing. Although CRISPR/Cas9 is all the rage, they had another reason to test this system. It’s difficult to determine how much protein iTOP causes a cell to take up, and continued protein expression would require multiple transduction events. For these reasons, iTOP is best suited to binary systems in which transient protein expression has a long-lasting effect, such as Cre- or Cas9-mediated recombination.
Both primary hESCs and immortalized cells treated with iTOP efficiently took up both Cas9 protein and a gRNA (yes, iTOP can also be used for RNA transduction) targeting a gene associated with diphtheria toxin lethality and were sorted into single wells to generate clonal populations. After iTOP treatment, both primary (hESCs) and cultured cells showed up to 70% resistance to diphtheria toxin, indicating a 70% frequency of biallelic mutations. Importantly, hESCs retained their stemness, indicating that iTOP may be valuable for CRISPR/Cas9 stem cell editing.
 |
| The small molecule NSDB and Na+ hypertonicity are key elements of iTOP. iTOP can be used to transduce Cas9 and a gRNA, which subsequently enter the nucleus and cleave the specified target. Figure adapted from D’Astolfo et al. |
In certain instances, the potential benefits of nonviral transduction are numerous. First, you remove the risk of viral integration causing mutations and gene expression changes elsewhere in the genome. Second, expression is by nature transient, which may lower nuclease toxicity or the potential for off-target effects. Third, protein purification (once a protocol has been established) may be less laborious than viral purification, and it does not raise the safety concerns associated with virus work. Fourth, working directly with proteins removes the limitations of packaging size associated with viral vectors. For these reasons, iTOP, especially in tandem with genome editing, may make primary cell work much more efficient and feasible for many labs.
References
-
Efficient Intracellular Delivery of Native Proteins. D'Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N. Cell. 2015 Apr 23;161(3):674-690. doi:10.1016/j.cell.2015.03.028. PubMed.
Find the Plasmids @Addgene:
Previous Protein Delivery Techniques:
-
Introduction of macromolecules into cultured mammalian cells by osmotic lysis of pinocytic vesicles. Okada CY, Rechsteiner M. Cell. 1982 May;29(1):33-41. PubMed.
-
Protein transduction: unrestricted delivery into all cells? Schwarze SR, Hruska KA, Dowdy SF. Trends Cell Biol. 2000 Jul;10(7):290-5. PubMed.
-
Is VP22 nuclear homing an artifact? Lundberg M, Johansson M. Nat Biotechnol. 2001 Aug;19(8):713-4. PubMed.
Topics: CRISPR, CRISPR Expression Systems and Delivery Methods






Leave a Comment