 This post was contributed by guest blogger Nathaniel Roquet, a PhD student in the Harvard Biophysics program and researcher in the Lu Lab at MIT.
This post was contributed by guest blogger Nathaniel Roquet, a PhD student in the Harvard Biophysics program and researcher in the Lu Lab at MIT.
Note: The following blog post reduces the content of our paper, “Synthetic recombinase-based state machines in living cells” (1), into a more straight-forward, concise explanation of how to adapt our engineered devices, recombinase-based state machines for your own experimental needs. For more context, exposition, and detail, please refer to the paper.
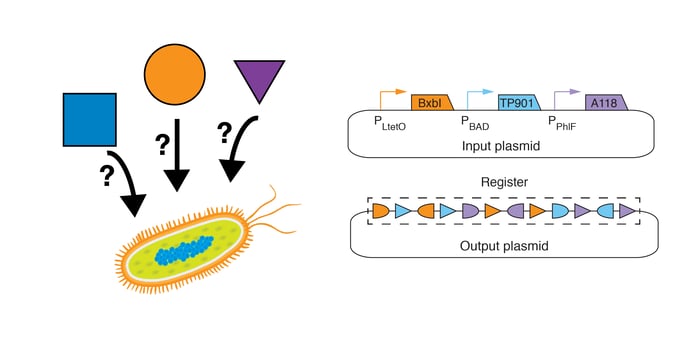
Why might one be interested in state machine technology?
Biological research has produced a massive amount of information regarding which regulatory proteins, signaling molecules, mutations, and environmental conditions drive certain cellular behaviors, but little is known about the order or timing of these factors. Recombinase-based state machines (RSMs), which take on a particular DNA-sequence configuration (state) based on the identity and order of a particular set of inputs, may be used to better understand and engineer cellular processes that are influenced by temporally ordered biochemical events.
How do RSMs work?
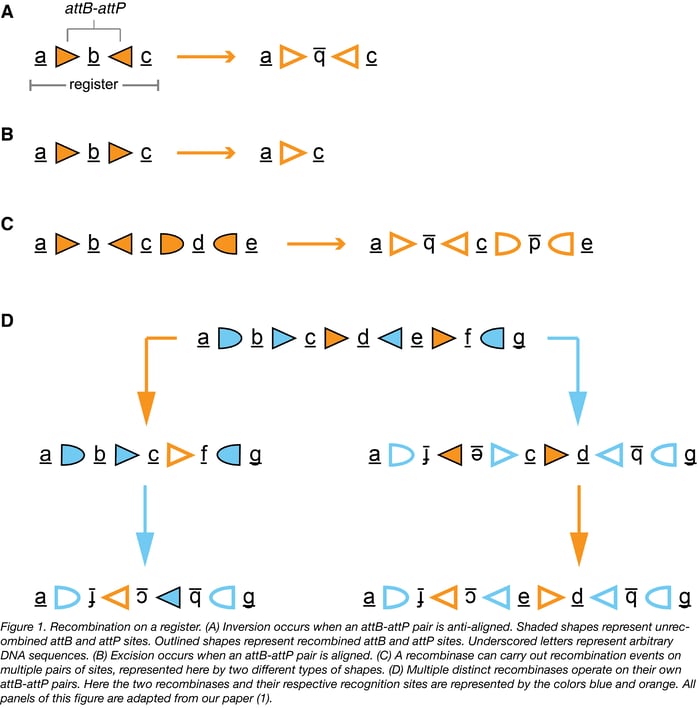
The state of an RSM is defined by the sequence within a prescribed region of DNA termed the “register”. Each chemical input to an RSM drives expression of a distinct large serine recombinase that recognizes its own attB and attP recognition sites on the register and catalyzes their recombination, thereby changing the state of the RSM. If an attB-attP pair is aligned (sites facing the same direction) on a register, then the result of their recombination is the excision of the DNA between the sites (Fig. 1B below). Alternatively, if an attB-attP pair is anti-aligned (sites facing opposite directions) on a register, then the result of their recombination is the inversion of the DNA between them (Fig. 1A below). In the absence of extra recombinase-specific co-factors (which should be excluded from the system), these reactions are irreversible.
Once expression of a particular recombinase is triggered, its effects on the state cannot be undone and also limit the set of possible downstream states that can be achieved by expression of other recombinases. Multiple orthogonal attB-attP pairs per recombinase may be arranged on a register (Fig. 1C below). If attB-attP pairs from different recombinases are overlapping or nested on a register, then the operation of one recombinase on the register could affect the way downstream recombinases interact with the register- either by re-orienting their recognition sites, or by excising one or multiple of their recognition sites from the register. If the RSM is designed in such a way that each possible order of inputs results in only one final state, then the state of the RSM will tell you precisely what inputs were added to the system and in what order they were added. Figure 1D below demonstrates an example of a register that enters distinct states when treated with different combinations and orders of two recombinases.
How might one read the state of an RSM?
The most straightforward way to read the state of an RSM is to sequence the register. In our paper (1), we allowed individual bacterial cells to form colonies on a plate and then we performed colony PCR on the register region and sent the PCR product for sequencing.
A less straightforward, but more population-wide (as opposed to single-cell) approach is to interrogate state with qPCR. The re-arrangement of DNA segments in each state of a register enables the design of primer pairs that PCR amplify in some states but not others. Because samples of many registers will likely not be homogenous for one state, regular PCR may not be very informative – each primer pair will amplify even if its target states are only represented in a very small number of registers. Instead, one may use qPCR to determine the fraction of registers that are amplified by a particular primer pair. Then, by leveraging qPCR data from multiple primer pairs, one can infer the predominant state of registers within a sample using a metric that we describe in the Materials and Methods section of our paper (1).
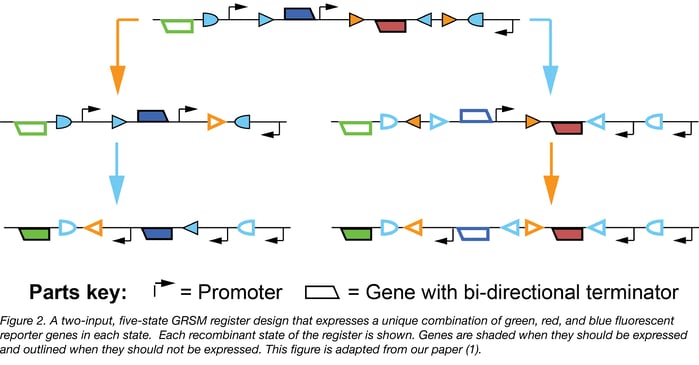
Lastly, it is possible to build gene regulatory RSMs (GRSMs, the subset of RSMs that regulate gene expression) that uniquely express selectable and/or sortable reporters in certain states of interest. For example, in Figure 2 below we present a two-input, five-state GRSM register that expresses a unique combination of red, blue, and green fluorescent proteins in each state.
The three methods of state interrogation mentioned above (Sanger sequencing, qPCR, and fluorescent reporter-based) each come with their advantages and disadvantages. Sanger sequencing is simple and the most accurate method as it directly reads the state of a register. However, each Sanger sequencing reaction can only be performed on one register at a time. If a particular application makes it difficult to isolate single registers or if it requires reading so many registers that Sanger sequencing would be too expensive, then the qPCR method should be considered as an alternative. The qPCR method can be performed on a sample of multiple registers. However, it is only semi-quantitative. It won’t tell you the distribution of states in a sample; instead, it will suggest the predominant state of registers in a sample based on a similarity metric that we defined in our paper. In our opinion, the most convenient strategy is the fluorescent reporter based method since it lends itself to cheap, quick, and high throughput state detection with flow cytometry. However, expressing reporters will consume cellular resources and therefore this state read-out method may not be suitable for applications that require minimal cellular perturbation. Also, this method requires live, intact cells; the sequencing and qPCR methods do not.
How might one implement RSMs?
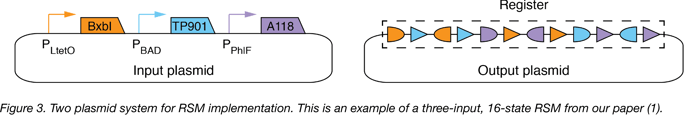
In our study (1), we implement RSMs in E. coli with two plasmids (Figure 3 below):
- Input Plasmid: The input plasmid (high copy, ColE1 origin of replication), contains large serine recombinase genes downstream of inducible promoters: the BxbI gene downstream of the anhydrotetracycline (ATc)-inducible promoter (PLtetO), the TP901 gene downstream of the arabinose (Ara)-inducible promoter (PBAD), and the A118 gene downstream of the diacetylphloroglucinol (DAPG)-inducible promoter (PPhlF).
- Output Plasmid: The output plasmid (single copy, bacterial artificial chromosome backbone) contains the register.
Our paper (1) explains how to arrange attB-attP pairs on a register to record all permuted substrings of a set of chemicals. Specific examples of arrangements that should be able to record all permuted substrings of up to seven chemical inputs are given in Table S2 of our paper. The paper also demonstrates that genetic parts (genes, terminators, and promoters) may be interweaved onto a GRSM register to execute predictable gene regulation programs that specify which genes should be expressed or not expressed in each state. To help researchers adapt GRSMs for their research, we provide MATLAB-based software that automates the design of registers that can be used to execute user-specified two-input gene regulation programs. The software has two components - a database of pre-compiled register designs and a search function, both of which can be found on GitHub along with an instructional text file.
What's available at Addgene?
In total, we constructed and tested eight two-input RSMs and three three-input RSMs. The plasmids used to construct these RSMs are listed in the table below and can be requested through Addgene. Their plasmid maps and GenBank files can be found on their plasmid pages. All two-input GRSMs use the same recognition site array on the register, and all three-input GRSMs use the same recognition site array on the register.
| RSM (See performance data from the indicated figures in our paper (1)) | Input Plasmid | Output Plasmid |
| Fig. 3A | pNR64 | pNR160 |
| Fig. 4A | pNR220 | pNR188 |
| Fig. 6A | pNR64 | pNR163 |
| Fig. 6B | pNR64 | pNR186 |
| Fig. 6C | pNR64 | pNR165 |
| Fig. 6D | pNR64 | pNR164 |
| Fig. 6E | pNR64 | pNR291 |
| Fig. 7A | pNR220 | pNR292 |
| Fig. 7B | pNR220 | pNR284 |
| Fig. S10A | pNR64 | pNR166 |
| Fig. S10B | pNR64 | pNR187 |
To construct your own two-input or three-input GRSMs, we recommend using overlap extension PCR and Gibson assembly to piece together the registers. All parts used in this paper (recognition sites, terminators, promoters, and genes) were built with flanking primer binding sites (designed to have 20-30 nt length, ~60 oC annealing temperature, and ~50% GC content) so that individual parts and/or strings of parts may be PCR-ed out of their current registers and used to build new registers. The primer binding sites can also be used to design primers to read states by qPCR or Sanger sequencing as described in the Material and methods section of our paper (1).
How might one troubleshoot GSRMs that don't work as expected?
The successful implementation of GRSMs depends on the reliability of the parts used to build them. We recommend using strong, directional terminators and promoters with buffer sequences on their 5’ and 3’ ends (for insulation). A list of promoters and terminators used in our GRSM registers can be found in Table S4 of our paper (1). These parts are also annotated on the plasmid maps and GenBank files uploaded to Addgene. We also recommend inserting self-cleaving hammerhead ribozymes directly upstream of the RBS for any gene on a register. The purpose of this is to prevent interference between the transcribed gene and its 5’ UTR. Various self-cleaving hammerhead ribozyme parts are described in Nielsen et al (3) and Lou et al (4).
Despite choosing proper register parts and designs, we have observed that certain GRSMs may still not perform as expected, perhaps due to idiosyncrasies that arise when all parts (recognition sites, promoters, genes, terminators, etc) are composed. Fear not – the state machine design space is highly degenerate and often times there are multiple register designs that may be used to implement the same gene regulation program. We recommend testing multiple register designs for the same gene regulation program to find the one that best suits your application.
How efficiently do RSMs work?
We tested a two-input (ATc and Ara) RSM over a time series of increasing input exposure lengths and found that two hour inductions were sufficient for greater than 50 percent of RSM-containing cells in a population to correctly record each possible permuted substring of the inputs, and that five hour inductions were sufficient for greater than 90 percent of cells to correctly record each possible permuted substring of inputs (see Figure S9 of our paper (1)).
We anticipate that, depending on the context and application, some inducible recombinase systems may work faster or more efficiently than others. Multiple large serine recombinases have been described in the literature (5, 6) and may be substituted for one another in an RSM. The predictability of RSMs and recombinase-based circuits in general would benefit from a more comprehensive characterization of the recombinases at our disposal.
Many thanks to our guest blogger Nathaniel Roquet!
 Nathaniel Roquet is a PhD student in the Harvard Biophysics program and researcher in the Lu Lab at MIT. He is broadly interested in biocomputing and biological circuits.
Nathaniel Roquet is a PhD student in the Harvard Biophysics program and researcher in the Lu Lab at MIT. He is broadly interested in biocomputing and biological circuits.
References
1. N. Roquet et al., Synthetic recombinase-based state machines in living cells. Science. 353, aad8559 (2016). PubMed PMID: 27463678.
2. Green, Alexander A., et al. "Toehold switches: de-novo-designed regulators of gene expression." Cell 159.4 (2014): 925-939. PubMed PMID: 25417166. PubMed Central PMCID: PMC4265554.
3. Nielsen, Alec AK, et al. "Genetic circuit design automation." Science352.6281 (2016): aac7341. PubMed PMID: 27034378.
4. Lou, Chunbo, et al. "Ribozyme-based insulator parts buffer synthetic circuits from genetic context." Nature biotechnology 30.11 (2012): 1137-1142. PubMed PMID: 23034349. PubMed Central PMCID: PMC3914141.
5. Brown, William RA, et al. "Serine recombinases as tools for genome engineering." Methods 53.4 (2011): 372-379. PubMed PMID: 21195181.
6. Yang, Lei, et al. "Permanent genetic memory with> 1-byte capacity." Nature methods 11.12 (2014): 1261-1266. PubMed PMID: 25344638. PubMed Central PMCID: PMC4245323.
Additional Resources on the Addgene Blog
- Learn How Piggy Bac Vectors Can Be Used for Genome Editing
- Learn about Michael Koeris' Journey from Grad Student to SynBio Entrepeneur
- Read All of Our SynBio Posts
Additional Resources on Addgene.org
- Visit Our SynBio Pages
- Check Out More Lu Lab Plasmids
- Find More Genome Engineering Tools
Topics: Synthetic Biology, Other Plasmid Tools, Plasmids





Leave a Comment